September 2024
This release note includes:
- Updates to the eProtein Discovery Cloud Software
- Updates to the eProtein Discovery Screen Experiment (5.1.0)
New Workflow Release
eProtein Discovery™ Screen Experiment 5.1.0
This workflow will only be available on instruments that have been updated to software version 4.6.2. The following features will not be available retrospectively.
New Features
- A new Data Analytics tab has been added that displays charts of the experiment results.
eProtein Discovery™ (Cloud) Software Release
New Features
-
Users now have the capability to bulk import protein sequences from a FASTA file by clicking on the + New Protein icon. This enhancement is particularly beneficial for users looking to import 24 sequences or more at once.
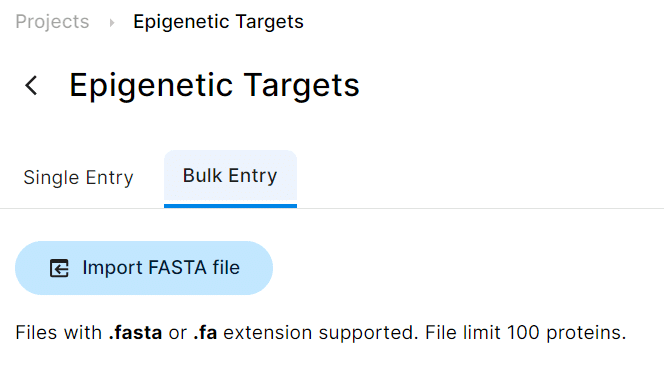
-
New status icons have been introduced in the Protein tab, allowing users to quickly view and check run status of each protein without needing to navigate to individual protein pages for details.
-
Experiments can now be duplicated and renamed.
-
Improved validation of DNA synthesis complexity:
- Complexity score with IDT recommendation will be displayed in full.
- A warning will be presented if DNA synthesis complexity is above a threshold of 10.
- High complexity amino acid sequences are blocked from proceeding to the next step.
-
Protein sequences can now be exported in CSV/FASTA format.
-
Improved eGene normalization and sequence calculation in the Possible Constructs tab.
-
Addition of support links, bold, italics, etc., to notes fields in proteins, projects, and experiments.
-
Amino acid sequences for all possible constructs are now available in the Possible Constructs table.
-
Protein sequences can now be exported in CSV or FASTA format by clicking on the ellipsis at the end of the protein list.
Bug Fixes
- Several bug fixes to improve user experience.