eProtein Discovery™ System - Standalone
eProtein Discovery system
📄 Download Page as PDFGeneral information
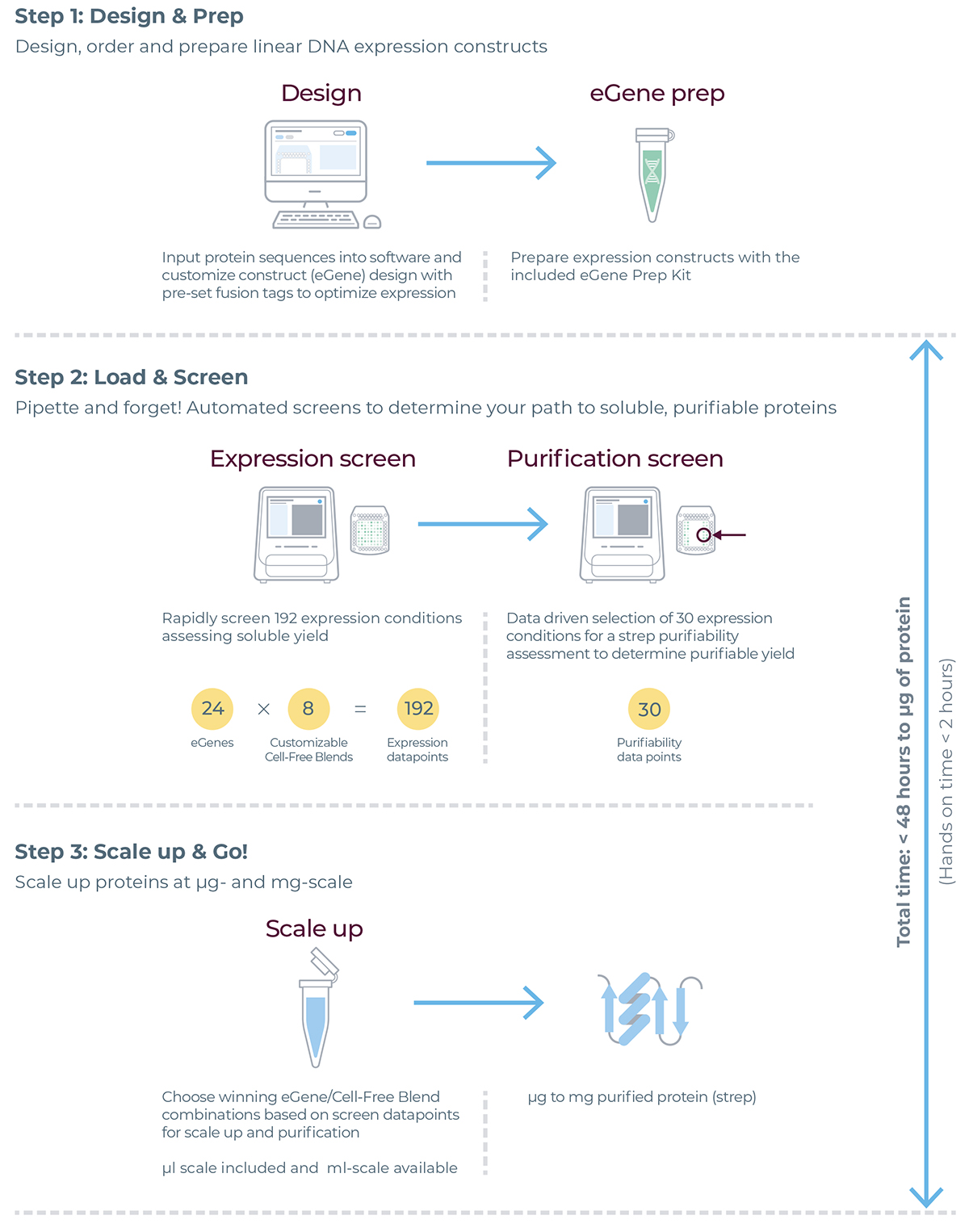
eProtein Discovery™ is the only end-to-end protein screening system that accelerates construct design, expression, solubility characterization and purification of target proteins in drug discovery programs. Accelerating the journey to your protein.
▷Rapid protein screening accelerates scientific progress by allowing researchers to quickly determine which proteins and their variants are optimal for achieving soluble, high yield proteins
▷Simultaneously screen multiple constructs and protein synthesis reagents for soluble expression, and then scale up to micrograms of recombinant protein off cartridge to test in your applications.
▷Explore multiple DNA constructs, including solubility tags, polymorphisms and isoforms on the same cartridge to expand your range of accessible proteins.
Four system components. One complete protein solution.
Instrument:
With protein prototyping, you can draw a straight line from theory to reality, allowing you to test hypotheses more efficiently and focus on promising targets. The eProtein Discovery™ instrument puts rapid protein prototyping on your benchtop.
Designed for all levels of scientist, it streamlines your workflow and grants you the ability to identify optimal DNA constructs, test expression feasibility earlier, and pursue targets with confidence. Fail fast, succeed faster!
Software: eProtein Discovery™ software simplifies a complex multivariate experimental design. The software sets up and simultaneously tracks 192 different combinations of DNA sequences, flank pairs and expression reagent reactions performed on eProtein Discovery™ system. AI performs highly rigorous QA checks during an experiment to ensure data quality and consistency.
Cartridge:
Powered by digital microfluidic technology, software controlled digital signals guide the movement of droplets on the eProtein Discovery™ Cartridge surface to enable splitting, dispensing and merging of biological reagents. Pipette DNA, cell free expression reagents and purification solutions on the Cartridge and the technology will orchestrate the rest.
Gain precise control of your eGene™ constructs and reagents to screen and discover optimal expressing conditions within 24 hours, accelerating target selection. A simple set-up allows anyone to run the system with minimal training.
Reagents:
The reagents within the eProtein Discovery™ system allow you to optimize protein obtainability by characterizing and purifying different combinations of DNA constructs and expression conditions. Our system will screen 192 different combinations in 24 hours for you to select the optimal conditions to scale up and get protein.
Our eProtein Discovery™ software will guide you in creating the panel of DNA constructs and reagents to power your experiment. Our complete reagent package includes design and ordering of DNA, simplifying your workflow.
eProtein Discovery Workflow
eProtein Discovery product contents
Equipment
| Description | Quantity | Storage Temperature | Product Code | |
|---|---|---|---|---|
| eProtein Discovery Instrument | 1 unit | Room Temperature | N1001 |  |
Cartridge Kit NC3006 - Consumables
| Description | Quantity | Storage Temperature | Product Code | |
|---|---|---|---|---|
| eProtein Discovery Cartridge | 1 unit | Room Temperature | NC3006 |  |
| eProtein Discovery Cartridge Cover | 1 unit | Room Temperature | NC3012 | |
| Base Fluid | 1 unit | Room Temperature | NC3007 |  |
Cartridge Reagent Kit +4°C reagent - NC3010-2
| Description | Quantity | Storage Temperature | Product Code | |
|---|---|---|---|---|
| Strep Beads | 200 µL | +4°C | NC3010-2 |  |
Cartridge Reagent Kit -80°C reagents - NC3010-1 (yellow stripe on label)
| Description | Quantity | Storage Temperature | Product Code | |
|---|---|---|---|---|
| Cell Free Core Reagent | 160 µL | -80°C | SC3-01 |  |
| Blank Buffer | 150 µL | -80°C | SC3-02 | |
| Detector Protein* | 75 µL | -80°C | SC3-03 | |
| Universal Control* | 20 µL | -80°C | SC3-04 | |
| Complementation Control* | 20 µL | -80°C | SC3-05 | |
| Expression Control* | 20 µL | -80°C | SC3-06 | |
| Full Workflow Control* | 20 µL | -80°C | SC3-07 | |
| Wash Buffer* | 800 µL | -80°C | SC3-08 | |
| Elution Buffer* | 50 µL | -80°C | SC3-09 | |
| AdditiveBuffer* | 50 µL | -80°C | SC3-10 | |
| PDI/GSSG Mix* | 50 µL | -80°C | SC3-11 | |
| TRXB1* | 50 µL | -80°C | SC3-12 | |
| DNAk Mix* | 50 µL | -80°C | SC3-13 | |
| Zinc Chloride | 50 µL | -80°C | SC3-14 | |
| Calcium Chloride | 50 µL | -80°C | SC3-15 | |
| Manganese Chloride | 50 µL | -80°C | SC3-16 | |
| Cofactor Mix* | 50 µL | -80°C | SC3-17 | |
| GSSG* | 50 µL | -80°C | SC3-18 | |
| 3C protease* | 50 µL | -80°C | SC3-19 |
* Single use reagent that cannot be freeze/thawed multiple times.
User supplied reagents
- 5 nM eGene constructs (DNA), stored at -80°C, generated using the Nuclera eGene Prep kit NC3008 or NC3009
User supplied equipment
- Magnetic particle separator (compatible with 1.5 mL microcentrifuge tubes)
- Vortexer
- Microcentrifuge
- 1.5 mL microcentrifuge tubes
- 2-20 µL 8-channel pipette
- 2-20 µL single-channel pipette
- 200 µL compatible tips
Protein Variant Creation
The purpose of this guide is to describe a guided approach for designing protein variants, mutants, and truncated sequences.
Support users in generating variants of their protein to test on the eProtein Discovery platform and increase their chances to get quickly soluble, functional protein to use for downstream applications in their project.
Summary - A stepwise guided method for variant creation
▷ Step 1 - Identify Relevant UniProt ID Use sequence alignment (POI sequence) or direct UniProtID input to identify the starting protein sequence and/or several close protein family members - for example isoforms and splice variants. Annotate each starting sequence with all required metadata.
▷ Step 2 - Select Candidates Filter isoforms, align them and flag functional or structural domains of interest.
▷ Step 3 - Rule-based Sequence Editing I Apply simple rule based editing for each input Candidate. Depending on the domains present, each input Candidate sequence should generate several “virtual” constructs. Remove signal peptides and propeptides, it is also often beneficial to remove transmembrane domains (TMD).
▷ Step 4 - Rule Based Sequence Editing II - Terminal truncations Apply simple rule based editing for each input Candidate. Consider modifications around functional domains of interest, for example removing disordered or unnecessary domains. eProtein Discovery™ System User Guide 9
▷ Step 5 - Check for other known stable domains (NMR, X-Ray) Identify other important regions and create relevant variants.
▷ Step 6 - Compile final list of variant Candidates for a POI
A general guideline for manually designing gene fragments compatible with the eGene Prep Kit is available upon request, provided a Non-Disclosure Agreement (NDA) is in place. If you are unable to use the eProtein Discovery Cloud Software to design gene fragments and need further assistance with constructing fragments compatible with the eGene Prep Kit, please contact Technical Support. (techsupport@nuclera.com)
Details - A stepwise guided method for variant creation
| Step | Title | Input | Output | Operations |
|---|---|---|---|---|
| 1 | Identify Relevant Uniprot ID | Sequence or Uniprot ID | Annotated Uniprot sequences |
|
| 2 | Select Initial Candidates | Annotated Uniprot sequences | Isoforms and important domains flagged |
|
| 3 | Combine starting list | Seqs from steps 1 and 2 | List of input Candidates | Combine lists 1 and 2 |
| 4 | Rule-based SequenceE diting I - identify domains of interest | List of input Candidates | List of Child Candidates 1 Edited sequences named appropriately - rules applied see operations. A Child Candidate is a sequence derived from an Initial Candidate by applying Rule-based Editing - Step 4 |
|
| 5 | Rule Based Sequence Editing II - truncations | List of input candidates (Step 3) + List of Child Candidates (step 4) |
| |
| 6 | Compile final Screening Candidates | List of input candidates (Step 3) + List of Child Candidates (step 4) + List of Child Candidates (step 5) |
|
Additive Selection Guide
Depending on the protein of interest, the presence of additives may be needed to optimize the expression. We recommend that you review the feature, function and binder of each protein to help guide the selection of additives. These information can be found in Uniprot.
The decision tree in Figure 1 illustrates the Additive options based on protein requirements.
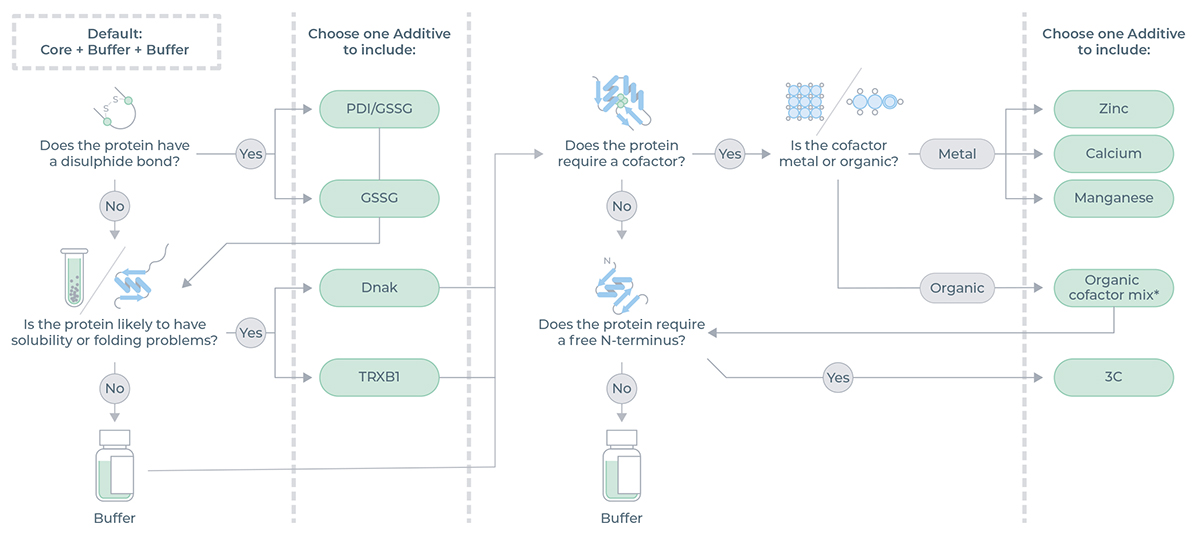 Figure 1: Decision tree which illustrates the Additive options based on protein requirements.
Figure 1: Decision tree which illustrates the Additive options based on protein requirements.
*Mix of NAD, acetyl-CoA, FAD, SAM and PLP.
The additives supplied in the Cartridge Reagent Kit NC3010 and their descriptions are listed in Table 1.
| Additive | Additive Description | Additive Characteristics |
|---|---|---|
| Additive buffer | HEPES buffer pH 7.5 and surfactant | CFPS reaction buffer, dilution normalization |
| PDI + GSSG Mix | Protein disulfide isomerase and oxidized glutathione | Chaperone and redox modification to oxidizing environment to support disulfide bond formation |
| TrxB1 | Thioredoxin reductase | Protects proteins from oxidative aggregation and inactivation and acts as a reductase in redox regulation |
| DnaK Mix | Chaperone | DnaK mix Chaperone mix to support folding and prevent aggregation |
| Zinc chloride | Zinc chloride solution | Cofactor that can be required for folding, stability, or activity |
| Calcium chloride | Calcium chloride solution | Cofactor that can be required for compaction, folding, stabilization, or activity |
| Manganese chloride | Manganese chloride solution | Cofactor for metalloenzymes for structure and activity |
| Cofactor Mix | Mix of NAD, acetyl CoA, FAD, SAM, and PLP | Cofactors that assist in folding, stability and activity |
| GSSG | Oxidized glutathione | Redox modification to oxidizing environment |
| 3C protease | 3C protease solution | Protease to cleave off the N-terminal solubility tag at the specific aminoacid sequence (LEVLFQ/GP) |
eProtein Discovery Software
Intended Use
The eProtein Discovery Software supports the user in the design and execution of combinatorial protein expression experiments on the eProtein Discovery platform.
Software Updates
▷ Software updates can be manually installed by connecting temporarily the instrument to the internet. Note: this applies if the instrument is able to connect to the eProtein Discovery Cloud Software.
Design an experiment
To design an experiment, use the Nuclera_eProtein_Discovery_Standalone_template (Excel file) available from the Nuclera Cloud Software, or on request by contacting the Nuclera Technical Support team (techsupport@nuclera.com).
The Nuclera_eProtein_Discovery_Standalone_template file is compatible with Microsoft Excel and it is not compatible with Google Sheet.
The excel template includes multiple worksheets. The worksheets provide step-by-step guide in designing the experiment and analyze data. Read the Template Guidance sheet before proceeding with experiment design.
Open the Excel file, enable editing, and save it under the name of your choice, for example the name or the date of the experiment.
"Template Guidance" sheet
This sheet contains a general introduction and guidance on how to use it. The first steps are to enable editing of the document and to save the file.
"1. Enter Experiment Details" sheet
In this sheet you will design your experiment by adding the combinations of proteins of interest (POI), solubility tags and additives selected for the experiment. Enter the requested information in the cells highlighted in yellow (Figure 2)
- Select the format of your experiment (3 proteins x 8 solubility tags, or 4x6 or 6x4 or 24x1)
- Enter the name of the proteins of interest
- Enter the molecular weights (kDa) for each protein of interest
- Select from the drop down menu the solubility tags chosen for the experiment
- Select from the drop down menus the two additives required for your experiment
The “Additive selection guide” section can help to select the right additives.
The eProtein Discovery™ system also provides the flexibility to incorporate custom additives into expression and purification workflows, enabling users to tailor conditions for unique protein targets. To ensure optimal performance and minimize risks, please consult the Compatibility List (https://info.nuclera.com/manual-custom-additives-chemical-compatibility-list.html), which provides detailed guidelines on compatible additives and their maximum allowed concentration. This resource serves as a valuable reference to help you achieve optimal results when working with custom additives.
Refer to this before experimenting with custom additives or contact Technical Support if you require more guidance.
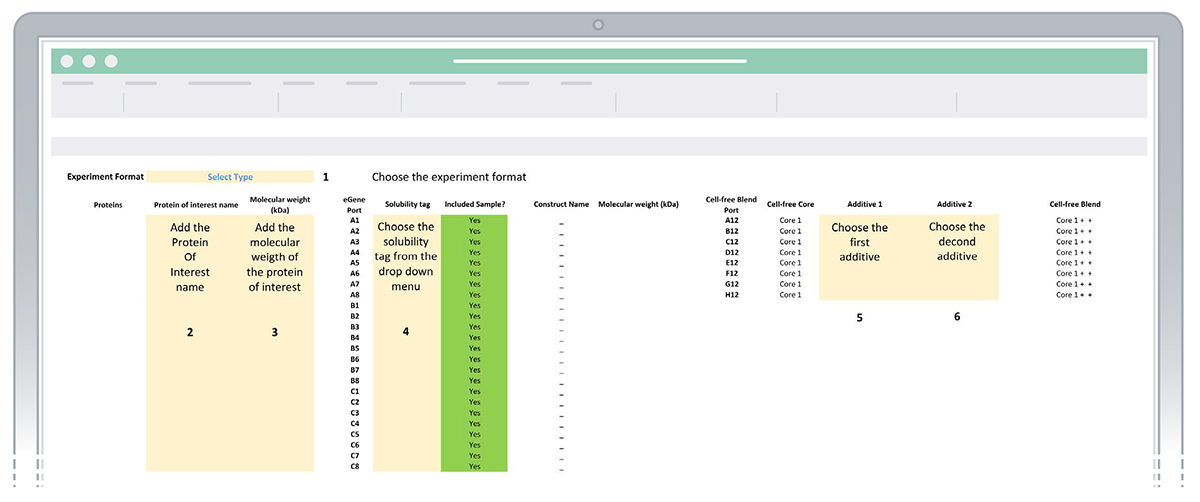 Figure 2: Sheet “1. Enter experiment details” of the eProtein Discovery Standalone template.
Figure 2: Sheet “1. Enter experiment details” of the eProtein Discovery Standalone template.
- Once the five steps outlined above are completed, the white columns for Construct, Molecular weight (KDa), Protein, and Cell-free Blend will be automatically populated with more information (Figure 3).
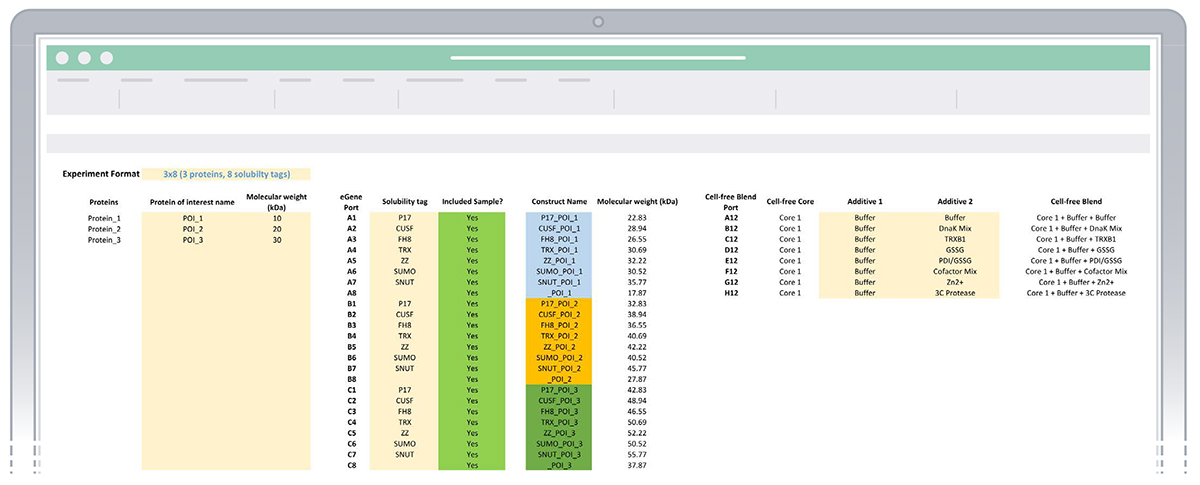 Figure 3: Example of sheet “1. Enter experiment details” with experiment details entered.
Figure 3: Example of sheet “1. Enter experiment details” with experiment details entered.
"2. Print plate map" sheet
This sheet is the printable version of the experiment design. It is recommended to print this sheet and take it to the lab as a guide for loading reagents onto the transfer plate.
- On the same sheet, below the table, you can find the transfer plate design (Figure 4). This design will help you determine where to load reagents on the transfer plate.
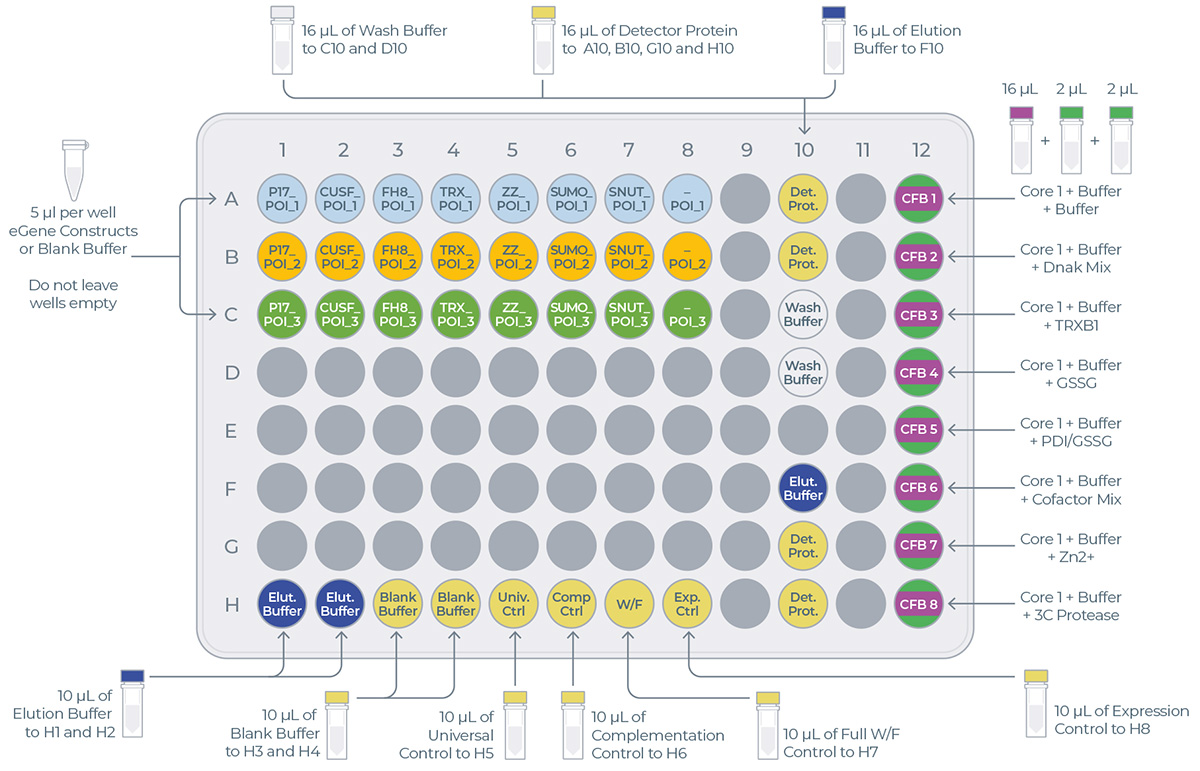 Figure 4: Example of transfer plate design
Figure 4: Example of transfer plate design
Preparation of the eProtein Discovery reagents
The preparation of the reagents takes about 1 hour.
Connect the vial of base fluid to the pump module
▷ In anticipation of starting a new experiment, take a fresh vial of base fluid, open it, and connect it to the left holder on the eProtein Discovery instrument pump module (Figures 5 and 6).
It is important to equilibrate the base fluid with the lab atmosphere prior to use. This is to prevent outgassing of the base fluid during the run, as air bubbles can interfere with the droplet movement. We recommend attaching the base fluid to the instrument the day before you will perform the run. An acceptable alternative is to incubate the uncapped base fluid at 30°C / 86°F for 1 hour.
▷ Connect an empty waste vial to the right holder of the pump module (Figures 6 and 7).
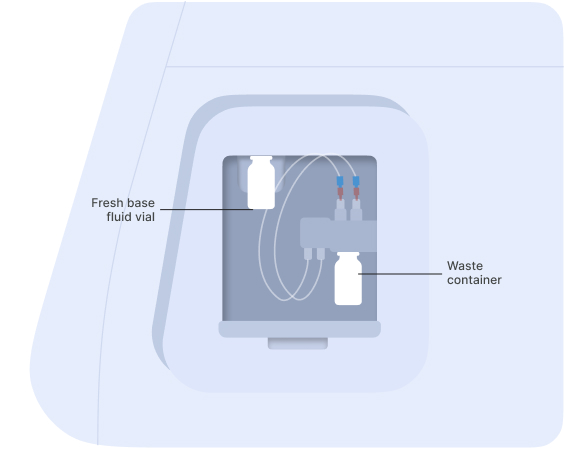
Figure 5: Vial of base fluid and the waste container connected to the pump as shown on the screen
[1]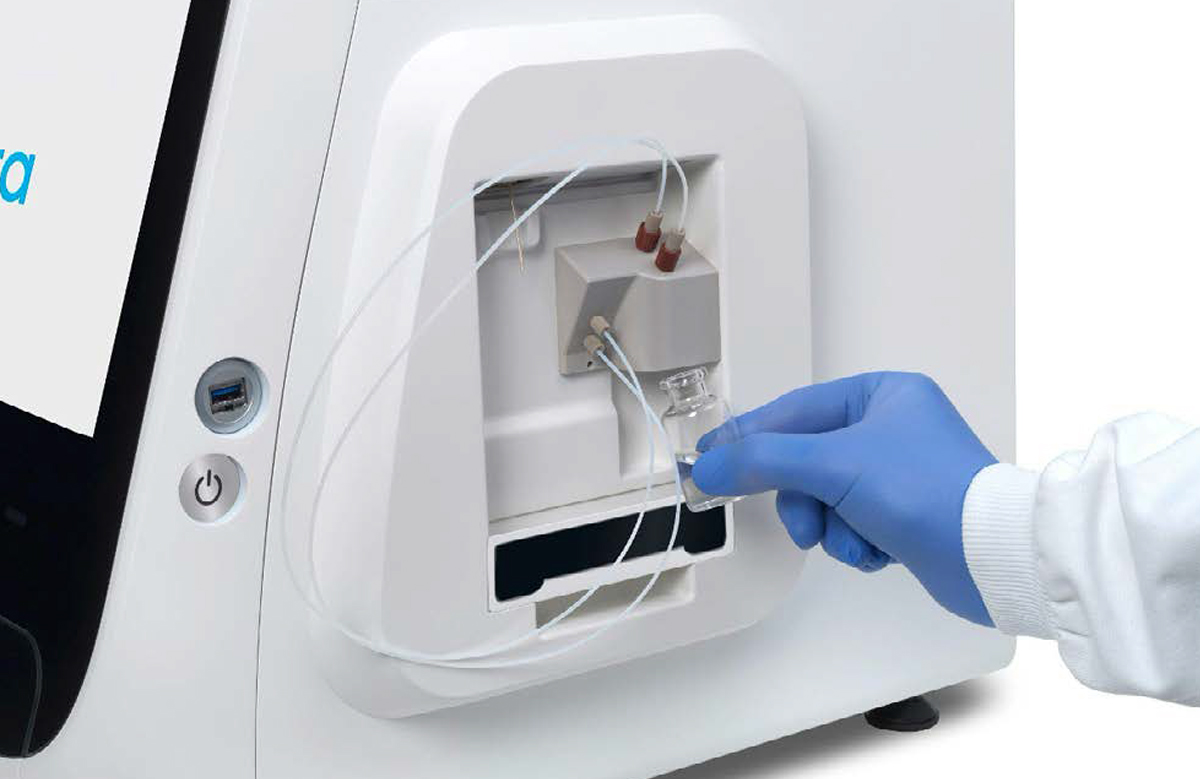 [2]
[2]
Figure 6: Connection of the empty waste container [1] and the vial of base fluid [2] to the pump
Prepare the transfer plate
After checking that the base fluid has been attached overnight to the instrument, take all the reagents out of the freezer.
The eProtein Discovery reagents need to be prepared and loaded onto a 96 well transfer plate following the layout and volumes in Figure 7 and Table 2.
it is critical to follow this layout exactly because it determines how the reagents are dispensed in the eProtein Discovery cartridge.
It is critical not to leave any port empty. If a eGene construct is missing it must be substituted with 5 µL eGene Elution Buffer supplied in the eGene Prep kit, not with water.
Tip: Empty ports can be used for duplicates.
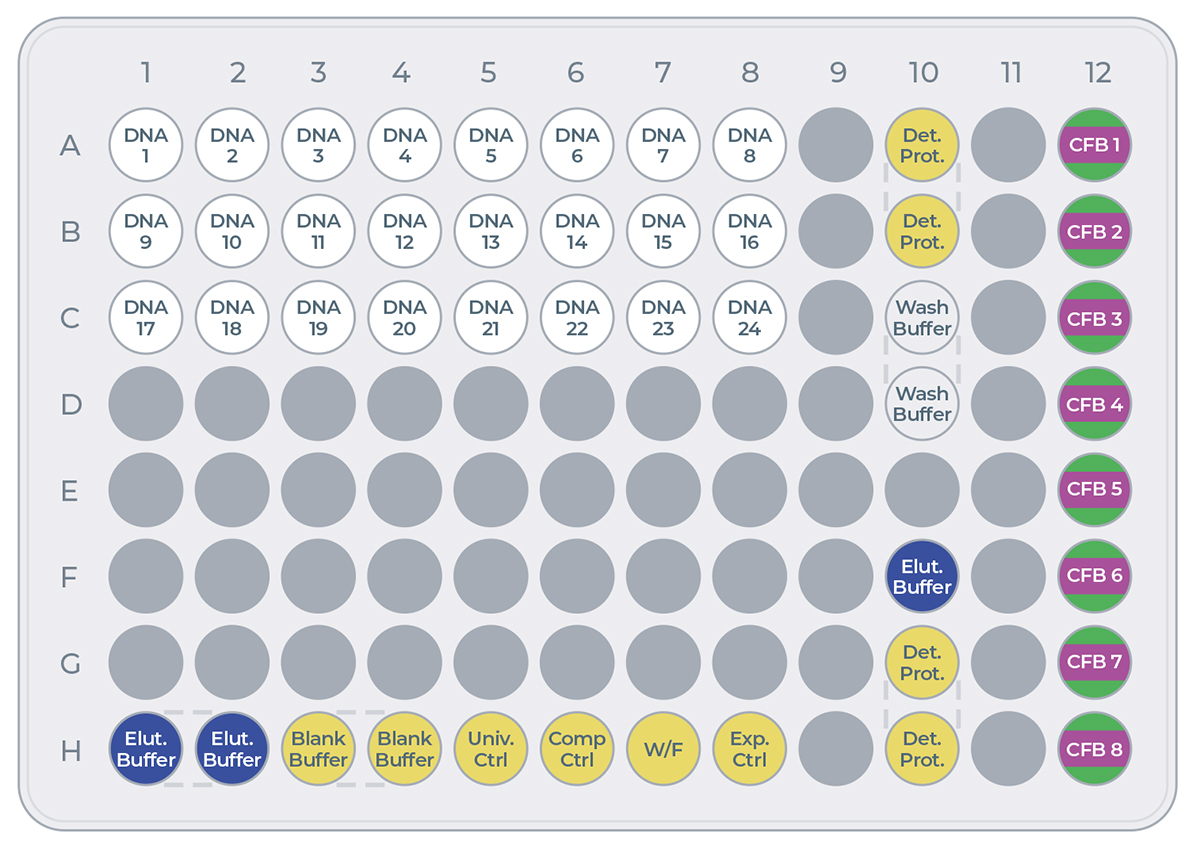 Figure 7: Transfer plate layout
Figure 7: Transfer plate layout
| Reagent | Volume (µL) |
|---|---|
| eGene construct | 5 |
| Controls: Blank Buffer, Universal Control (Univ. Ctrl), Complementation control (Comp. Ctrl), Full Workflow Control (W/F), Expression Control (Exp Ctrl) | 10 |
| Cell-free Blend (CFB): Cell-free Core Reagent + Additive 1 + Additive 2 | 20 (16+2+2) |
| Wash Buffer (Wash Buffer) | 16 |
| Elution Buffer (Elut. Buffer) | 10 µL in H1 & H2, 16 µL in F10 |
| Detector Protein (Det. Prot.) | 16 |
Table 2: Reagents and volumes to load on the transfer plate.
1. Take the Strep Beads from the fridge and the Cartridge Kit reagents (box with the yellow stripe on the label) from the -80°C freezer.

2. Place an empty 96-well transfer plate on ice.
Note: The transfer plate should be kept on ice until the transfer of reagents to the Cartridge.
Note: Ensure you prepare the Cell-free Blends last.
3. eGene constructs (DNA)
Take the vials or the plates with the eGene constructs made in advance using the eGene Prep Kit out of the freezer and thaw on the benchtop at room temperature. This takes approximately 15 minutes.
Note: the vials or the plates can be centrifuged for a few seconds to ensure all the liquid is at the bottom of the wells.
Load 5 µL of each eGene construct onto the transfer plate into wells:
▷ A1 to A8
▷ B1 to B8
▷ C1 to C8
Note: It is critical to load the eGene constructs onto the transfer plate in the exact order that they have been finalized in the experiment planned in the eProtein Discovery software.
4. eProtein Discovery purification reagents
Thaw the Wash Buffer and the Elution Buffer on the benchtop at room temperature. Once thawed, vortex for 2 seconds and centrifuge for 2 seconds using a microcentrifuge to mix and collect all the reagents.
▷ Load 16 µL of Wash Buffer into wells C10 and D10.
▷ Load 16 µL of Elution Buffer into well F10
▷ Load 10 µL of ELution Buffer into wells H1 and H2
5. eProtein Discovery controls
From the kit kept at -80°C, take the controls out and thaw them on ice.
▷ Load 10 µL of Blank Buffer into wells H3 and H4.
▷ Load 10 µL of Universal control into well H5.
▷ Load 10 µL of Complementation Control into well H6.
▷ Load 10 µL of Full W/F Control into well H7.
▷ Load 10 µL of Expression Control into well H8
6. Strep Purification Beads
Strep Purification Beads are provided in 200 µL aliquots of 5% v/v suspension – To prepare the Strep Beads:
- Take the vial of Strep Beads from the fridge and give it a quick spin for 2 seconds in a microcentrifuge to pellet the beads.
- Resuspend the beads by gently pipetting up and down 10 times with a p200 pipette set on 90 µL.
- Transfer 90µL of the resuspended beads into a 1.5 mL tube. Discard the rest only after the experiment starts, in case more volume is required.
- Place the tube with Strep Beads on a magnetic particle separator and capture for 1 min.
- Remove all the supernatant with a p200 pipette and discard the liquid.
- Remove the tube with Strep Beads from the magnetic particle separator. Resuspend the beads in 100 µL Wash Buffer by slowly pipetting up and down 10 times.
- Repeat steps 4 to 6 twice more for a total of three washes.
- After the third wash, spin down the tube and place it back on a magnetic particle separator and capture for 1 min.
- Remove all the supernatant with a p200 pipette and discard the liquid.
- Spin down the tube, place it back on a magnetic particle separator and remove the residual buffer with a p20 pipette.
- With a p20 pipette, resuspend the beads in 10.5 µL Wash Buffer by gently pipetting up and down 10 times to create a 15 µL 30% Strep Beads working
- Keep the beads in the tube on the bench, not on ice.
Note: The beads should NOT be loaded onto the transfer plate.
7. Detector Protein
The Detector Protein is supplied as ready to use. Spin down the tube for 2 seconds to collect the full volume at the bottom. Load 16 µL of Detector Protein into wells A10, B10, G10, and H10 of the transfer plate.
8. Preparation of the Cell-free Blends
For each expression screening experiment, up to eight 20 μL Cell-free Blends can be made by adding 16 μL of Cell-free Core Reagent, 2 μL of a first additive, and 2 μL of a second additive.
Note: The total volume of blend should always be 20 µL final
Note: the same additive can be used as first and second additive, for example 2 x 2 µL of Additive Buffer. The list of Additives is in Table 1.
- Thaw on ice Cell-free Core Reagents and Additives
- Once thawed, vortex for 2 seconds the Cell-free Core reagents and Additives to ensure they are well mixed.
- Centrifuge for 2 seconds the Cell-free Core reagents and Additives using a microcentrifuge to return any droplets to the bulk aliquot.
- Add 16 µL of Cell-free Core reagent to wells A12-H12.
- Add 2 µL of your first selected additive to wells A12-H12.
- Add 2 µL of your second selected additive to wells A12-H12.
Note: It is critical to load the Cell-free Blends onto the transfer plate in the exact order that they have been finalized in the experiment planned in the eProtein Discovery software.
Set up the experiment on the instrument
Press the [Power Switch] to activate the Instrument power-up, initialization and self-test sequence.
Create the experiment
- From the top right hand side of the screen select [Create Experiment]
- Select the 'eProtein Discovery Screen Experiment' workflow and press [Confirm] (Figure 8)
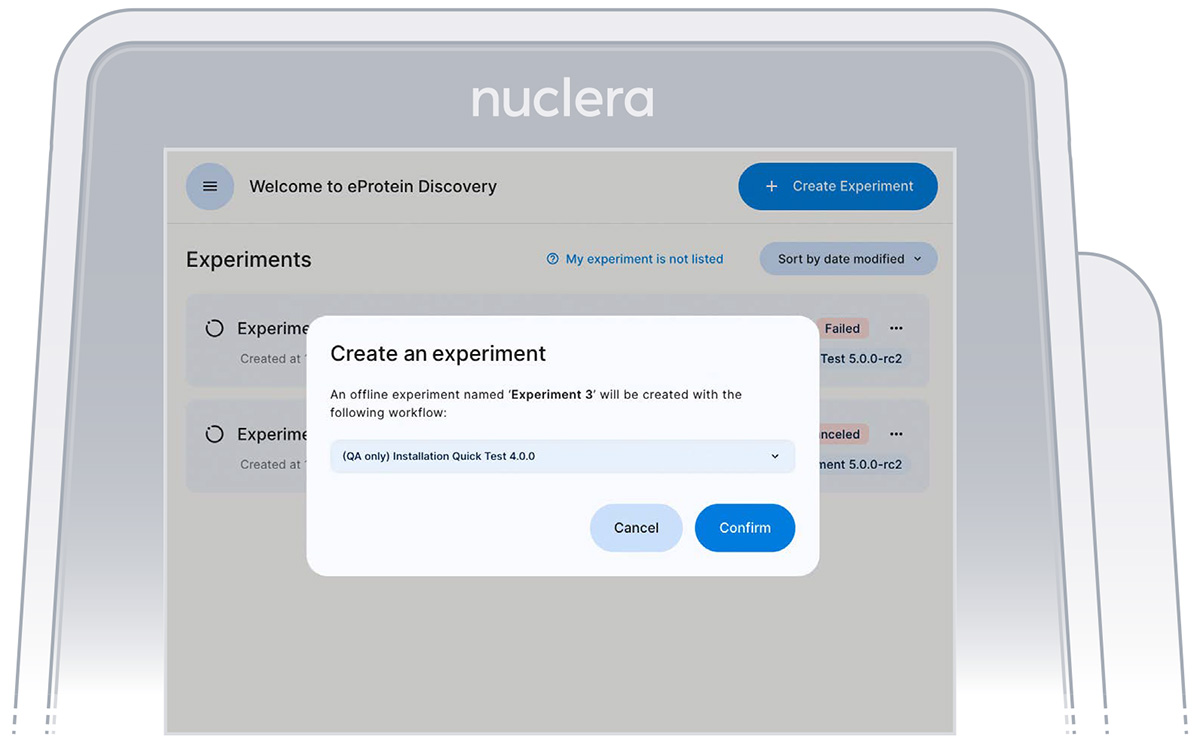 Figure 8: Experimental design screen
Figure 8: Experimental design screen
-
Select the type of experiment you would like to perform on the instrument (Figure 8). You can choose between:
a. 3x8 (3 proteins, 8 solubility tags)
b. 4x6 (4 proteins, 6 solubility tags)
c. 6x4 (6 proteins, 4 solubility tags)
d. 24x1 (24 proteins, FlexiVariant screen)
e. 30 highest expressing combinations
It is critical to select the exact experiment format as this will determine the downselection method of the 30 expression conditions for the strep purifiability assessment.
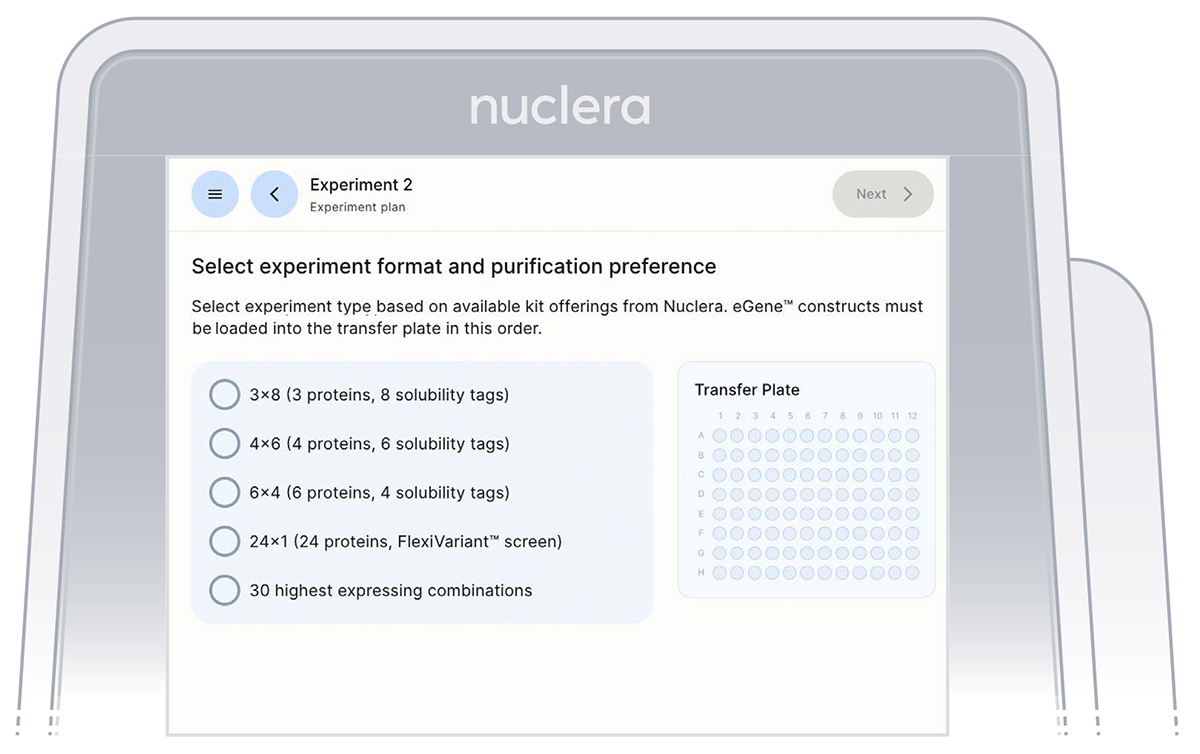 Figure 9: Instrument screen to select the experiment format
Figure 9: Instrument screen to select the experiment format
- Read the Before you proceed section and press the [Next] button (Figure 10).
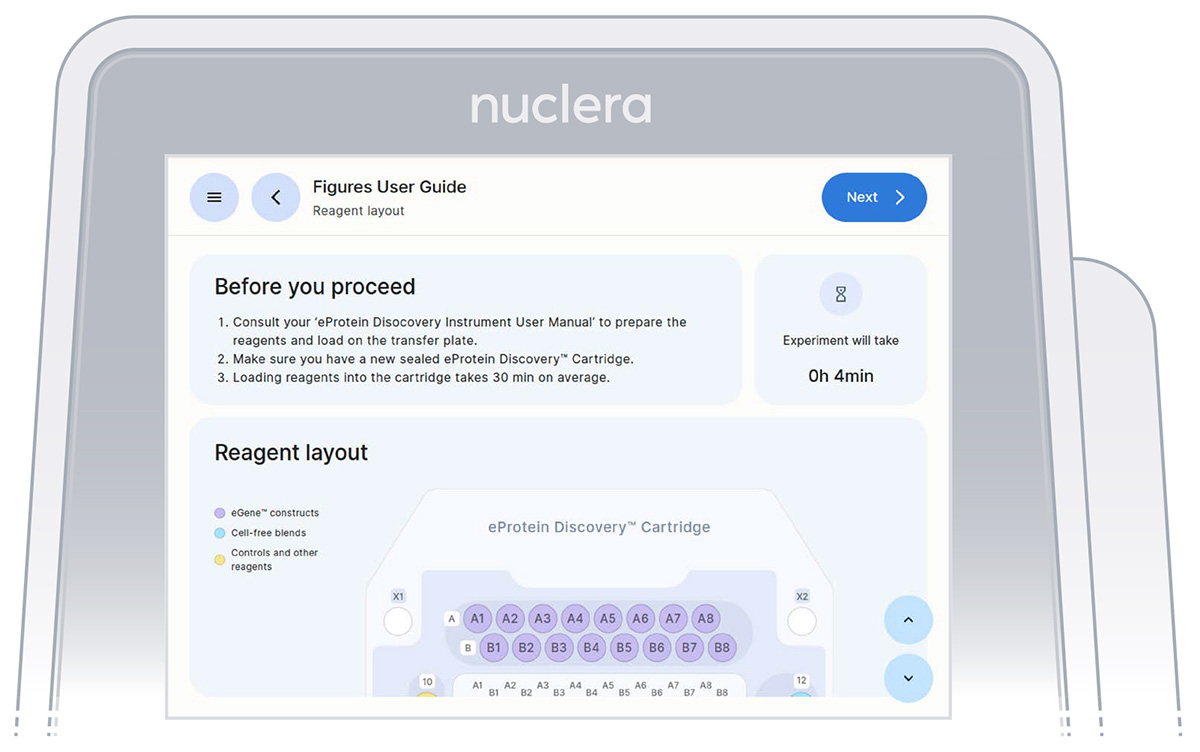 Figure 10: Summary of the selected experiment
Figure 10: Summary of the selected experiment
- Get the transfer plate containing the reagents and cartridge ready.
- Go through and tick the checklist, and press the [Next] button (Figure 11). The drawer will open.
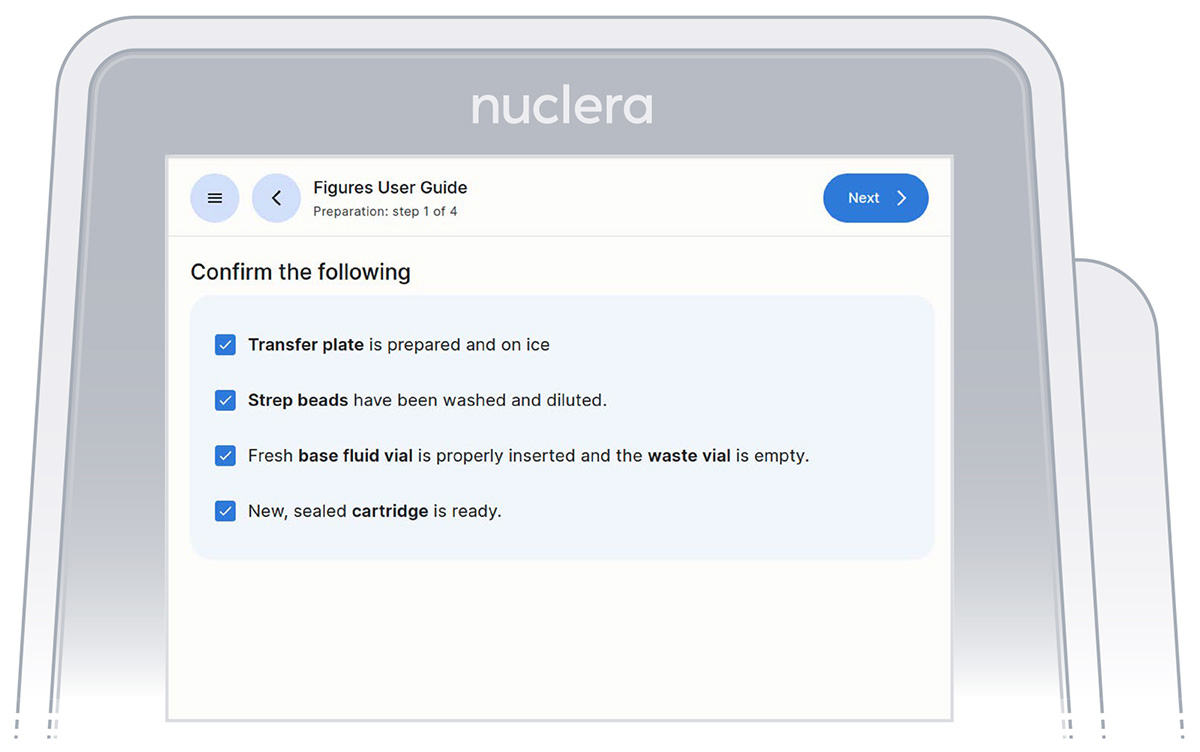 Figure 11: Checklist screen before the experiment starts
Figure 11: Checklist screen before the experiment starts
- Unpack and load a cartridge as shown on the screen of the eProtein Discovery instrument, place the cover on the cartridge, avoid touching the electrical connectors, and press the [Next] button (Figure 12).
Note: keep the cartridge packaging to dispose of the cartridge after use.
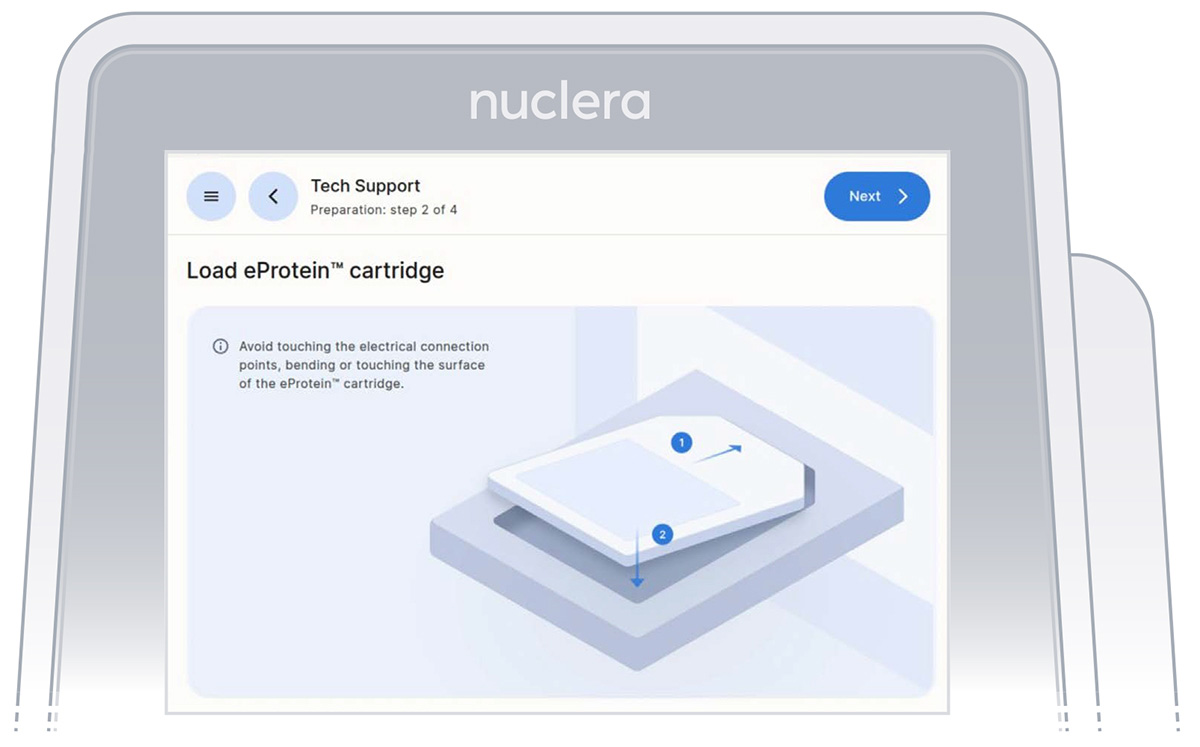 Figure 12: Loading of the cartridge on the eProtein Discovery instrument
Figure 12: Loading of the cartridge on the eProtein Discovery instrument
- Keep cover on the cartridge. Markings on the cover will guide you through the loading process.
Set up the pump on the instrument
Follow the on-screen instructions to complete the experiment.
▷ These instructions will guide you in operating the eProtein Discovery instrument and completing an experiment on the instrument.
▷ The instructions must be followed in the order shown on the screen.
▷ You can navigate forward and back through the steps using the buttons at the top.
▷ You can scroll up and down using the arrows at the bottom right of the screen when shown or with your fingers.
Note: once you start the experiment, the back button on the instrument will be disabled.
- On the right hand side of the instrument, ensure the tubings for the integrated pump are placed in the tubing holder, and press the [Next] button (Figure 13).
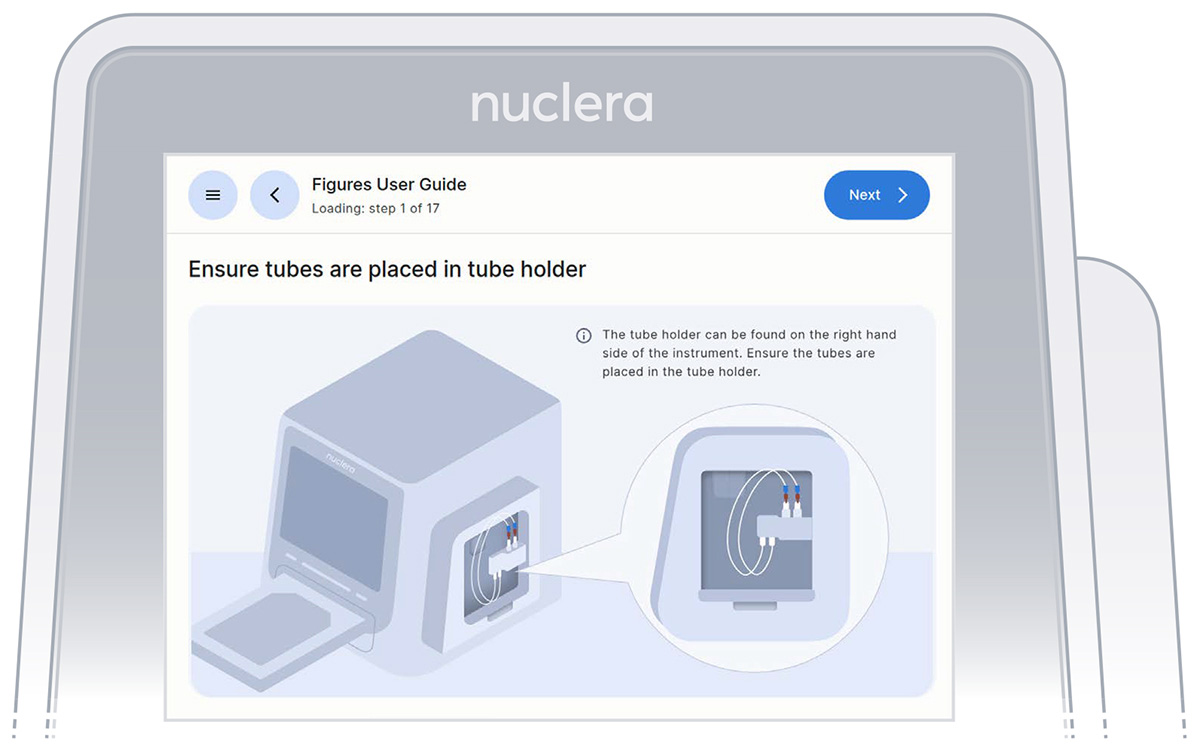 Figure 13: Loading of the cartridge on the eProtein Discovery instrument
Figure 13: Loading of the cartridge on the eProtein Discovery instrument
- Ensure the vial of base fluid and the waste container have been connected to the pump located on the right hand side of the instrument. Press the [Next] button (Figures 14).
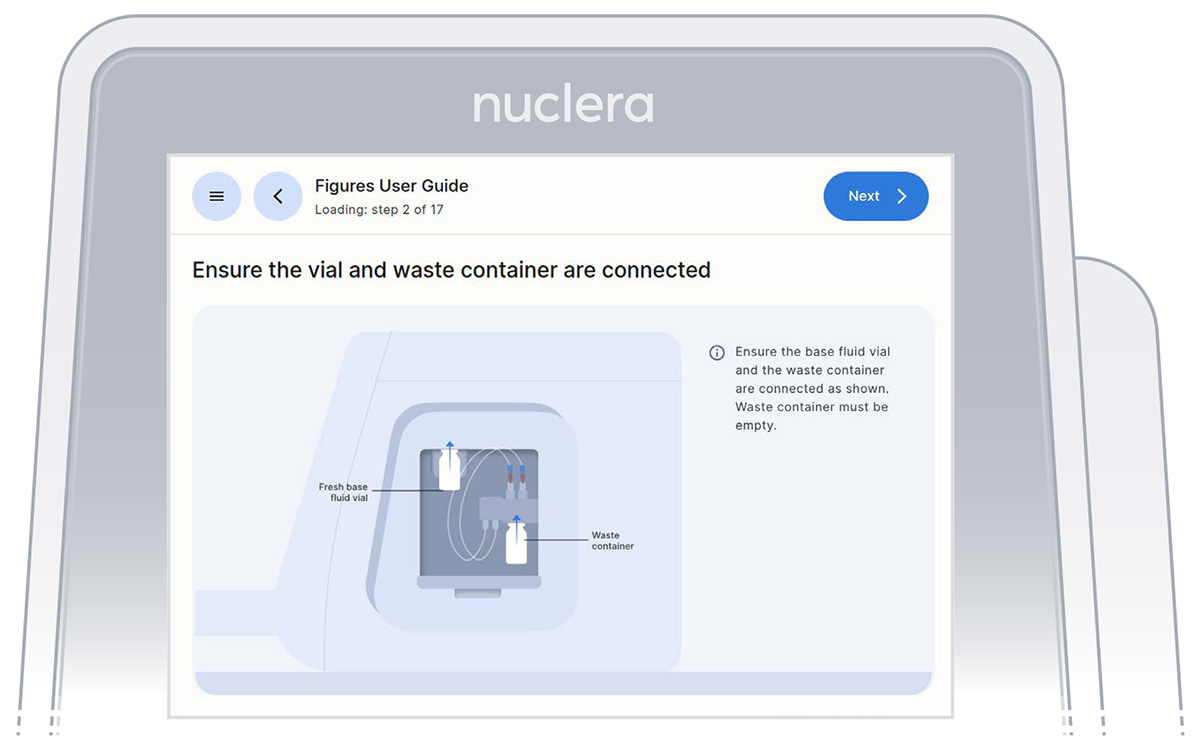 Figure 14: Vial of base fluid and the waste container connected to the pump as shown on the screen
Figure 14: Vial of base fluid and the waste container connected to the pump as shown on the screen
Filling the cartridge with base fluid
- With the tubing and containers in place, ensure that some of the base fluid has dripped into the waste container (Figure 15).
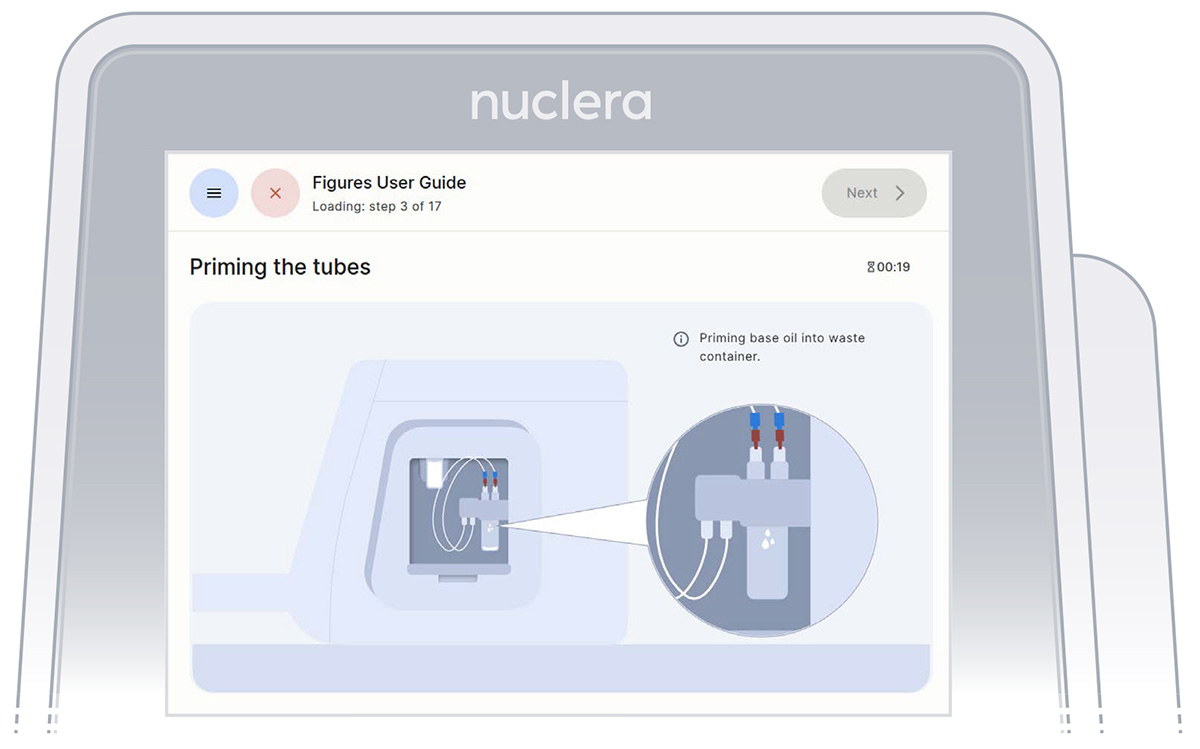 Figure 15: Priming the pump tubings with base fluid
Figure 15: Priming the pump tubings with base fluid
- Remove connectors tubing from the holder, connect them tightly to the corner ports X2 and X3 of the cartridge, and press the [Next] button (Figure 16). Either connector can be interchangeably inserted into corner port X2 or X3.
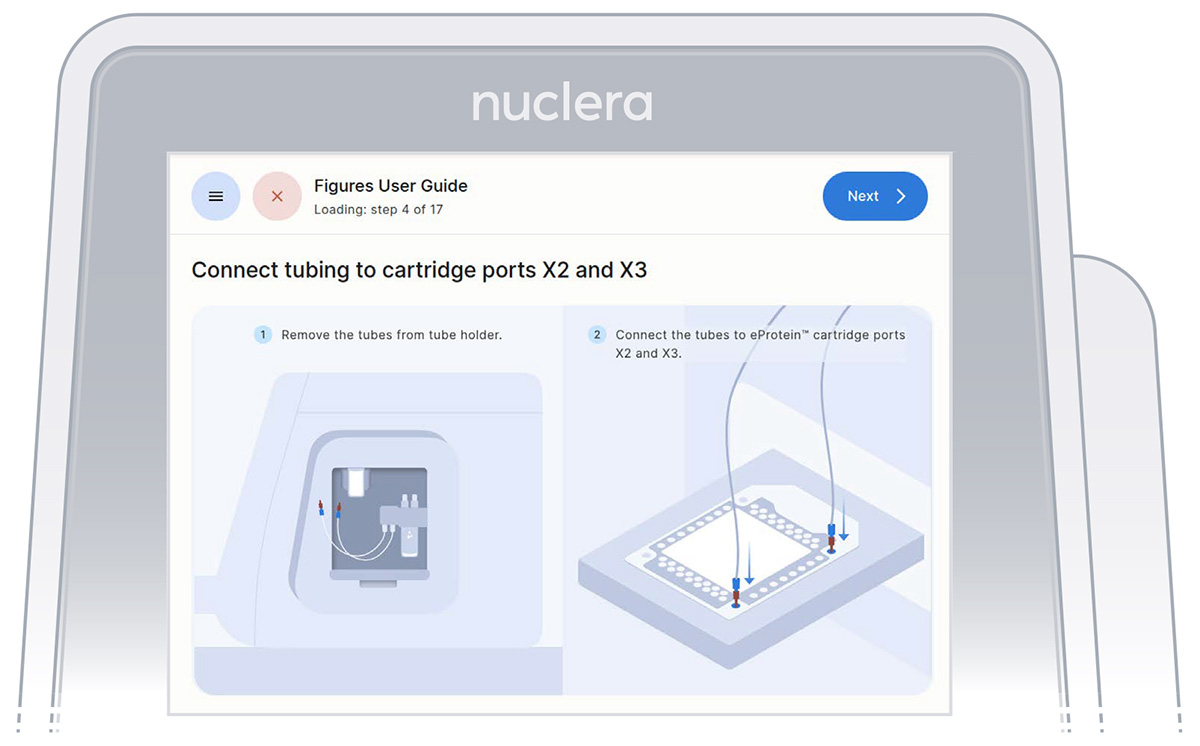 Figure 16: Connection of the pump tubes to the cartridge
Figure 16: Connection of the pump tubes to the cartridge
- Inspect the cartridge for air bubbles that may have been introduced during the priming with base fluid.
If any air bubbles persist after base fluid priming, use a single-channel p200 pipette to aspirate the air bubbles from the nearest port and reinject slowly the base fluid that was aspirated into a corner port (X1 or X4). Press the [Next] button (Figure 17).
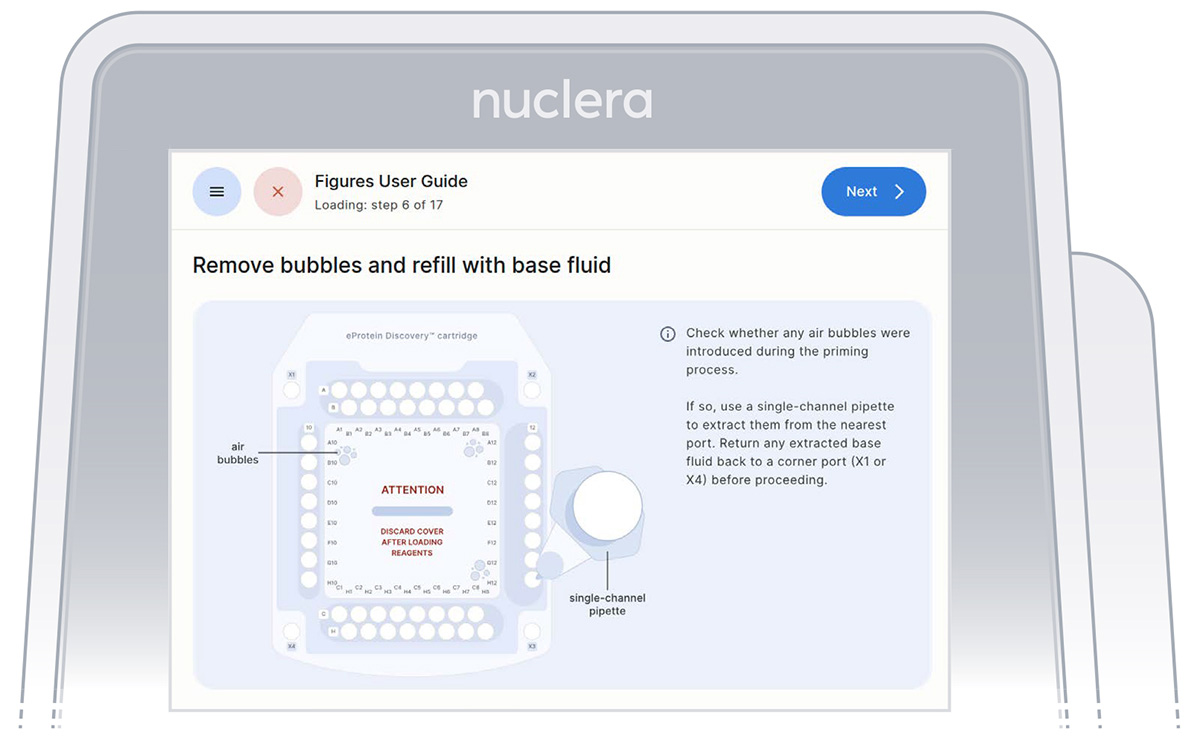 Figure 17: Procedure to remove potential air bubbles from the cartridge
Figure 17: Procedure to remove potential air bubbles from the cartridge
- Inspect the ports on the cartridge after the priming with base fluid is complete. Ensure all the ports are filled and press the [Next] button (Figure 18).
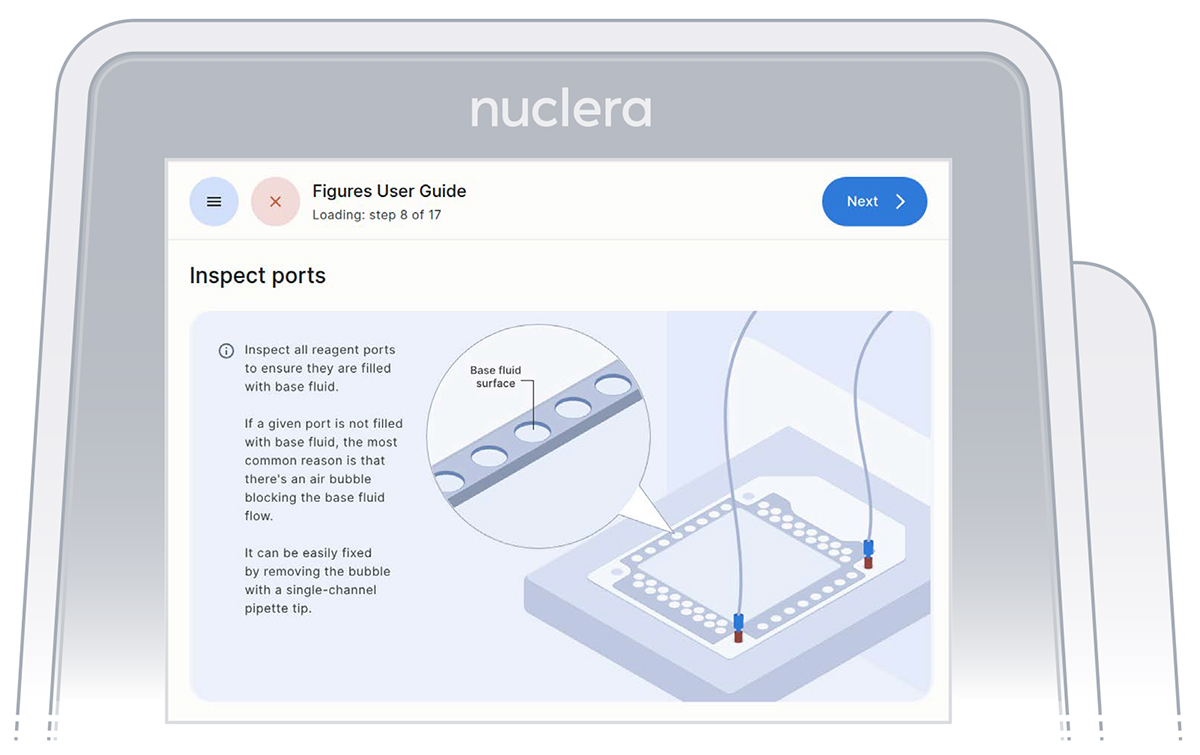 Figure 18: Procedure to remove potential air bubbles from the cartridge
Figure 18: Procedure to remove potential air bubbles from the cartridge
Load the reagents on the cartridge
Note: videos showing how to load the reagents on the cartridge can be found on the Nuclera website via this link: www.nuclera.com/resource-library/?resource_type=video.
Guidance for proper sample loading:
▷ Follow the on-screen instructions that will guide you in loading the reagents.
▷ The loading of the reagents should be done using a volume appropriate 8-channel pipette.
▷ To facilitate the pipetting of the reagents, the transfer plate can be moved from the ice bucket to the bench.
▷ Check the plate for the presence of air bubbles. Air bubbles can be removed by spinning the plate in a swing rotor centrifuge for about 10 seconds.
▷ After aspirating the reagents, make sure that all pipette tips are filled evenly, and contain no air bubbles.
▷ When loading the reagents into the ports, place the tip vertically in the port, just under the meniscus of base fluid. Do not touch the sides or base of the port.
▷ Dispense reagent gently to the first pipette stop. Do not go past the first stop.
▷ After dispensing, lift the tip while keeping the pipette plunger depressed. If using an electronic pipette, make sure that the purge function is disabled.
▷ Lift the tip while keeping the pipette plunger depressed.(Figure 19).
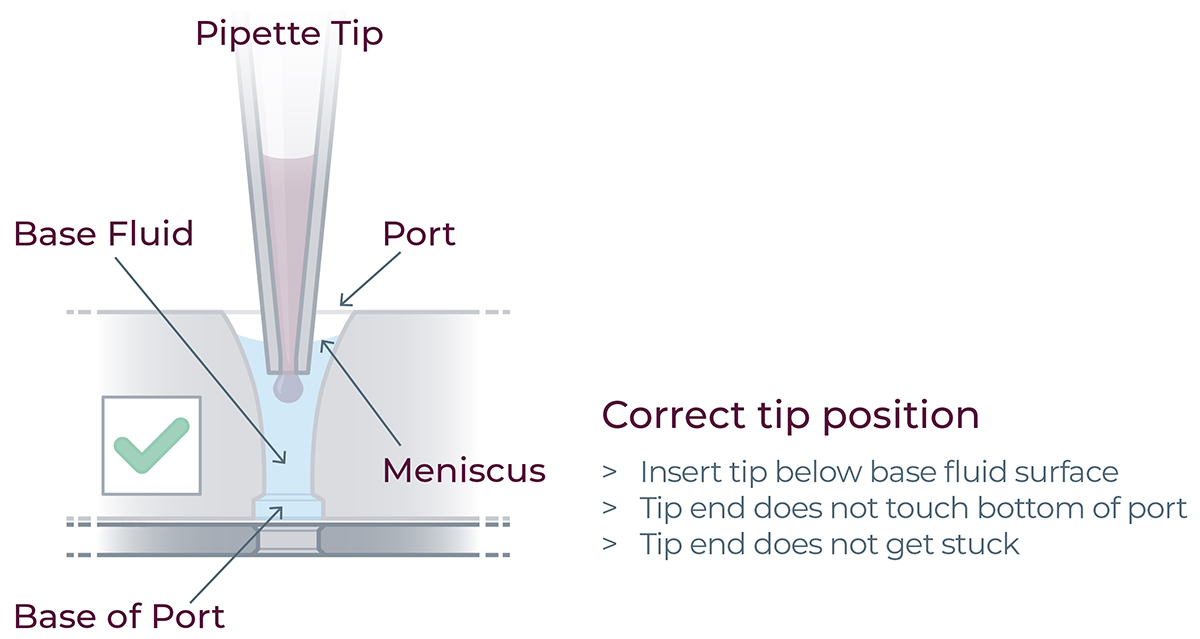 Figure 19: For correct reagent loading the pipette tip is immersed in the base fluid and not touching the bottom of the port
Figure 19: For correct reagent loading the pipette tip is immersed in the base fluid and not touching the bottom of the port
Electronic pipettes must be properly configured for use with eProtein Discovery. If you are using an electronic pipette, please contact Technical Support at techsupport@nuclera.com for assistance. Any run errors in eProtein Discovery caused by incorrect use of electronic pipettes are the responsibility of the user. We recommend using a manual multichannel pipette instead. Important settings to note for electronic pipettes are:
▷ Disable blowout/purge
▷ Avoid high speed for aspiration
▷ Avoid high speed for dispense.
Load eGene constructs - rows A, B and C
It is critical not to leave any port empty. If a eGene construct is missing it must be substituted with 5 µL eGene Elution Buffer supplied in the eGene Prep kit, not with water.
Tip: Empty ports can be used for duplicates.
▷ Load 8x fresh pipette tips and aspirate 3 μL of the eGene constructs from the transfer plate wells A1-A8 into ports A1-A8 of the cartridge (Figure 20).
▷ Load 8x fresh pipette tips and aspirate 3 μL of the eGene constructs from the transfer plate wells B1-B8 into ports B1-B8 of the cartridge (Figure 20).
▷ Load 8x fresh pipette tips and aspirate 3 μL of the eGene constructs from the transfer plate wells C1-C8
▷ into ports C1-C8 of the cartridge (Figure 20).
▷ Place the tip vertically in the port, just under the meniscus of base fluid and dispense slowly until the first stop of the pipette is reached. Do not engage the pipette tips fully into the ports.
▷ Eject the pipette tips into a waste container.
▷ Press the [Next] button on the screen.
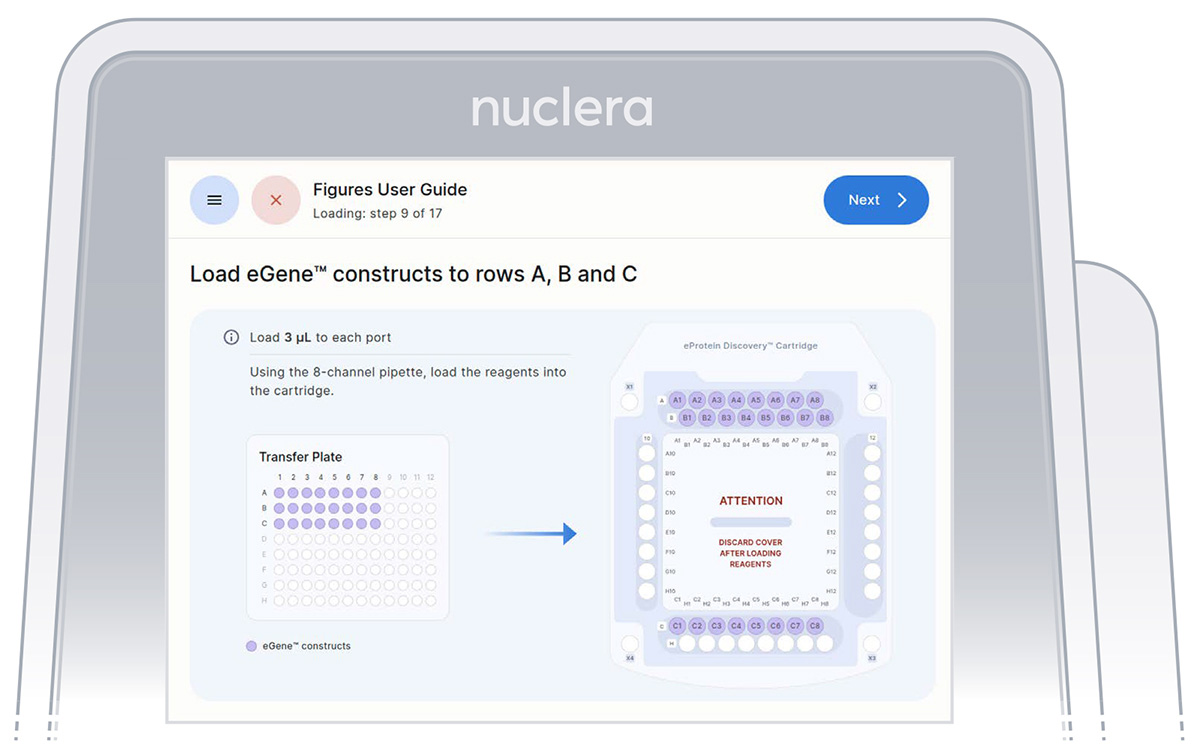 Figure 20: Loading of the eGene constructs onto row A, B and C of the cartridge
Figure 20: Loading of the eGene constructs onto row A, B and C of the cartridge
Load reagents - row H, column 12 and column 10
1. Reagents - row H:
▷ Load 8x fresh p20 pipette tips and aspirate 3 µL of the reagents from the transfer plate wells H1-H8 into ports H1-H8 of the cartridge (Figure 21).
▷ Place the tip vertically in the port, just under the meniscus of base fluid and dispense slowly until the first stop of the pipette is reached. Do not engage the pipette tips fully into the ports.
▷ Eject the pipette tips into a waste container.
▷ Press the [Next] button on the screen.
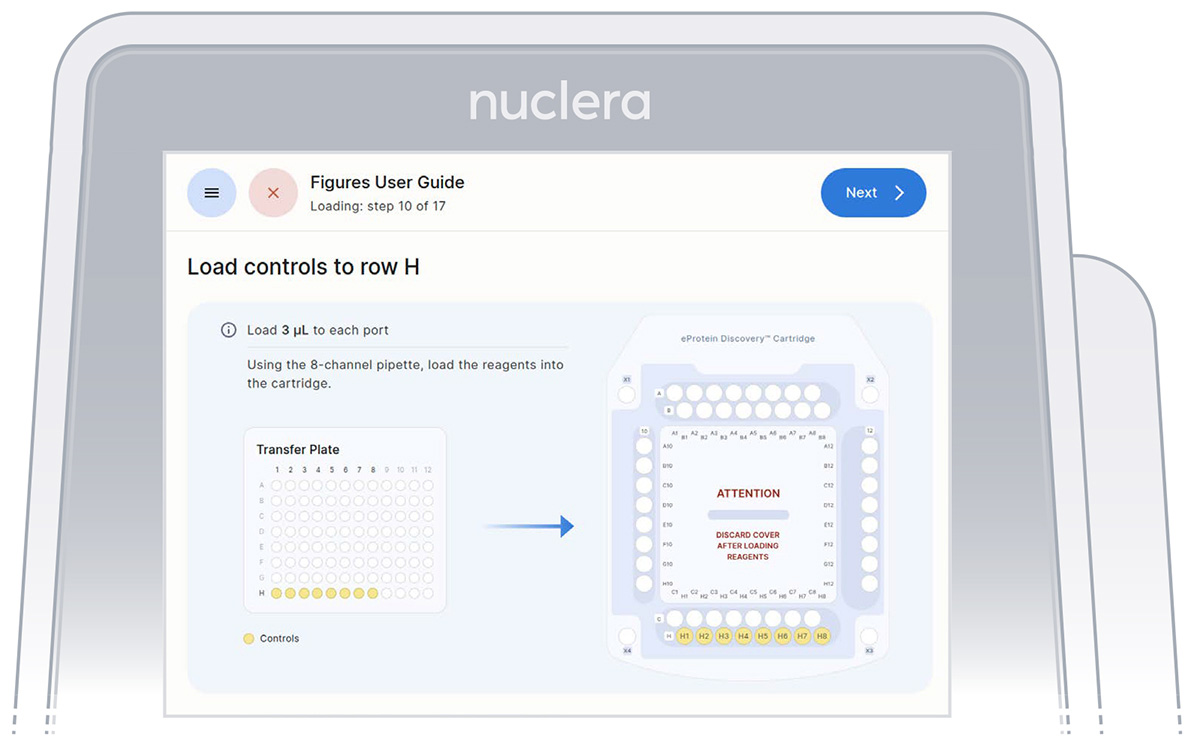 Figure 21: Loading of the reagents onto row H of the cartridge
Figure 21: Loading of the reagents onto row H of the cartridge
2. Reagents - column 12:
▷ Load 8x fresh p20 pipette tips and **mix the Cell-free Blends in the transfer plate by gently pipetting up and down 12 times. **
Be careful not to introduce air bubbles in the ports.
Aspirate 12 µL of the Cell-free Blends from the transfer plate wells A12-H12 into ports A12-H12 of the cartridge (Figure 22).
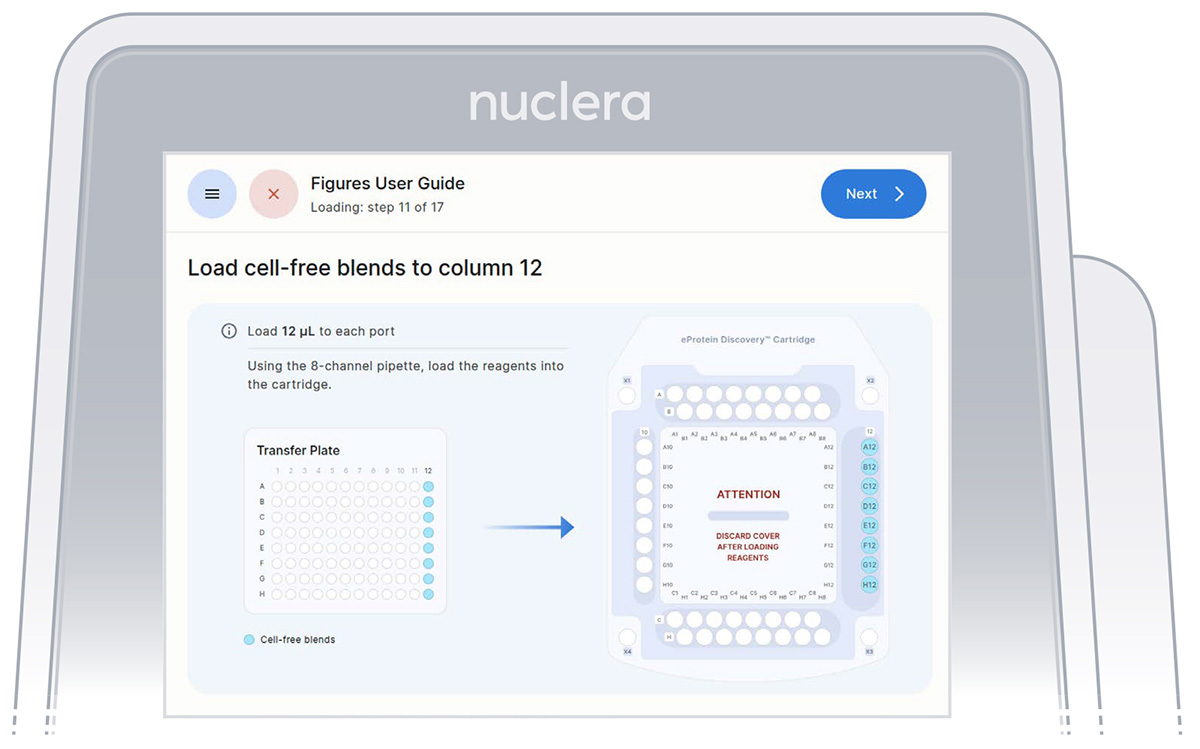 Figure 22: Loading of the Cell-free Blends onto column 12 of the cartridge
Figure 22: Loading of the Cell-free Blends onto column 12 of the cartridge
▷ Place the tip vertically in the port, just under the meniscus of base fluid and dispense slowly until the first stop of the pipette is reached. Do not engage the pipette tips fully into the ports.
▷ Eject the pipette tips into a waste container.
▷ Press the [Next] button on the screen.
3. Reagents - column 10:
▷ Load 7x fresh p20 pipette tips and aspirate 12 µL of the reagents from the transfer plate wells A10-D10 and F10-H10 into ports A10-D10 and F10-H10 of the cartridge (Figure 23).
▷ Ensure the tip is immersed in the base fluid and dispense slowly until the first stop of the pipette is reached. Do not engage the pipette tips fully into the ports.
▷ Eject the pipette tips into a waste container.
▷ Press the [Next] button on the screen.
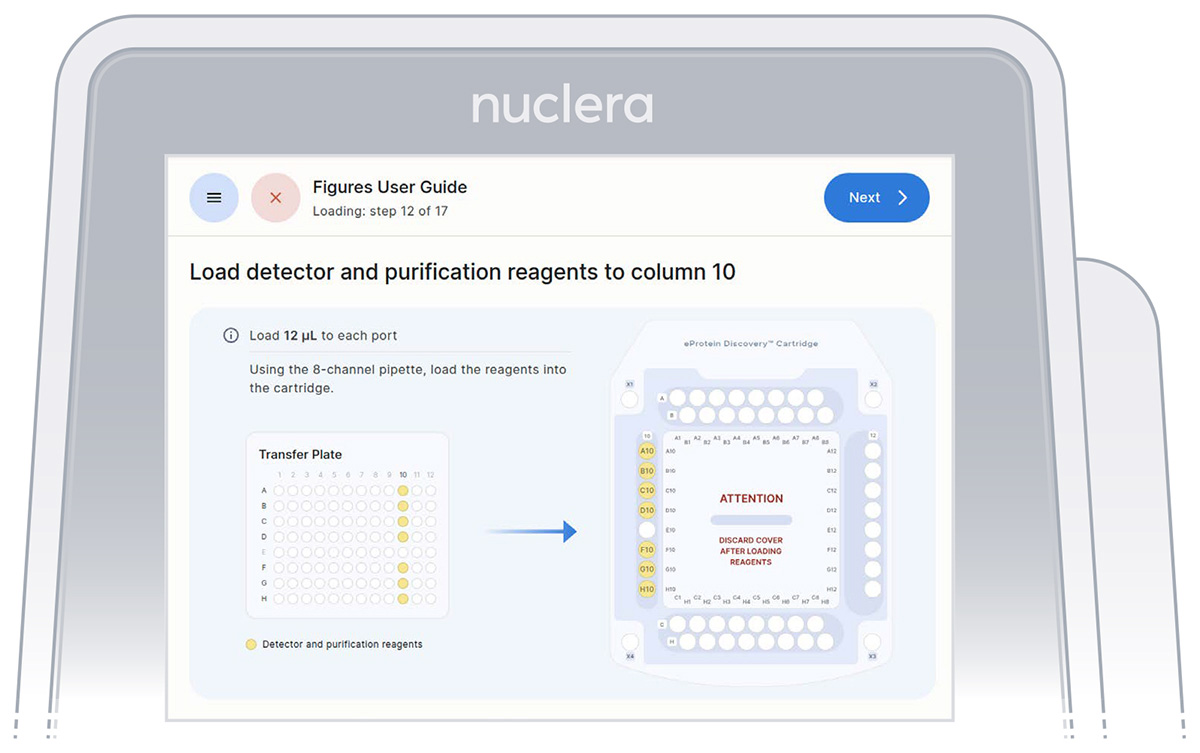
Figure 23: Loading of the reagents onto column 10 of the cartridge
4. Strep Purification Beads - port E10:
▷ Using a single channel p20 pipette, mix the Strep Purification Beads twelve times by gently pipetting up and down. Be careful not to introduce air bubbles. Aspirate 12 µL of the Strep Purification Beads prepared in a tube and dispense into port E10 of the cartridge (Figure 24).
▷ Place the tip vertically in the port, just under the meniscus of base fluid and dispense slowly until the first stop of the pipette is reached. Do not engage the pipette tip fully into the port.
▷ Eject the pipette tip into a waste container.
▷ Press the [Next] button on the screen.
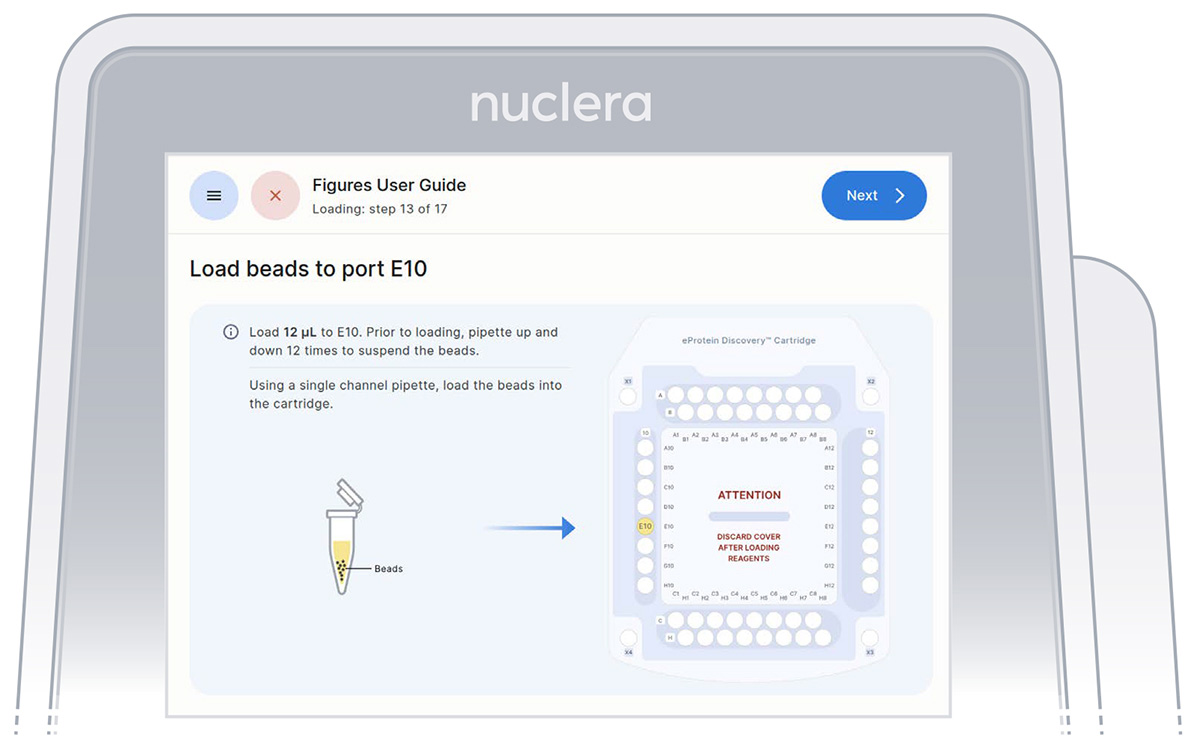 Figure 24: Loading of the Strep beads to port E10 of the cartridge
Figure 24: Loading of the Strep beads to port E10 of the cartridge
- Remove the cover from the cartridge (Figure 25)
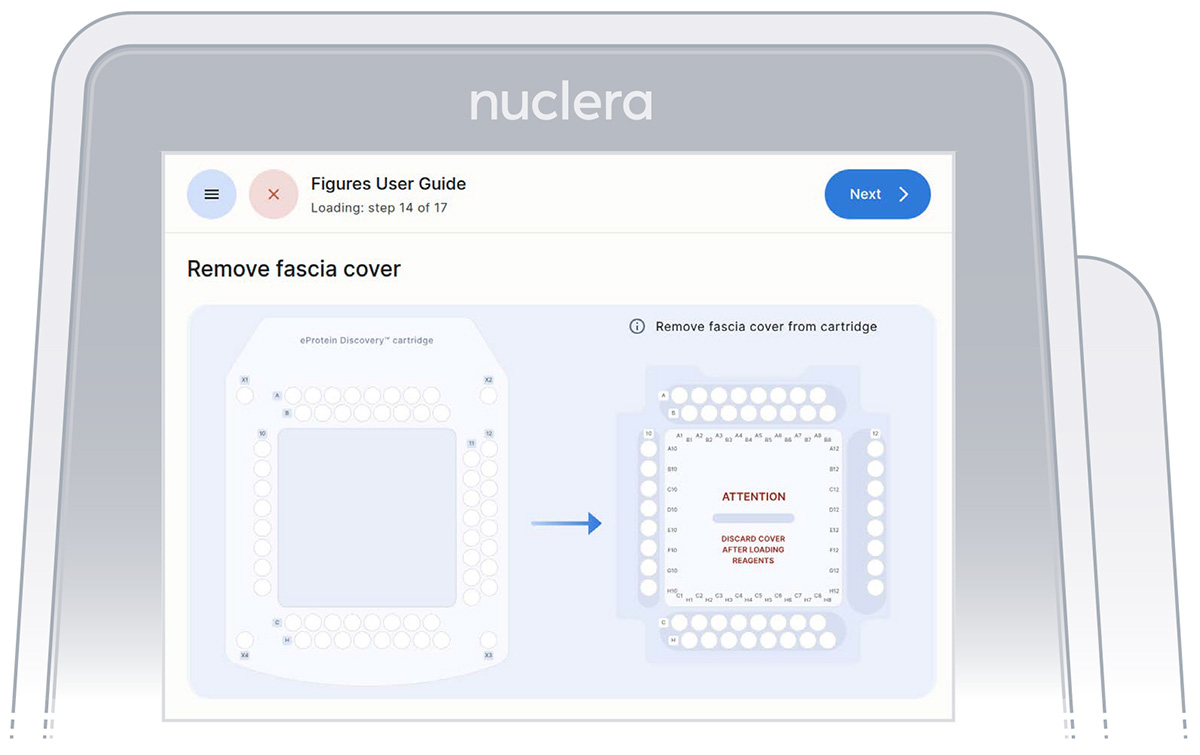 Figure 25: Remove the cover from the cartridge
Figure 25: Remove the cover from the cartridge
Load reagents in the cartridge
- Press the [Next] button to start the aspiration of the base fluid and the loading of the reagents on the cartridge (Figure 26).
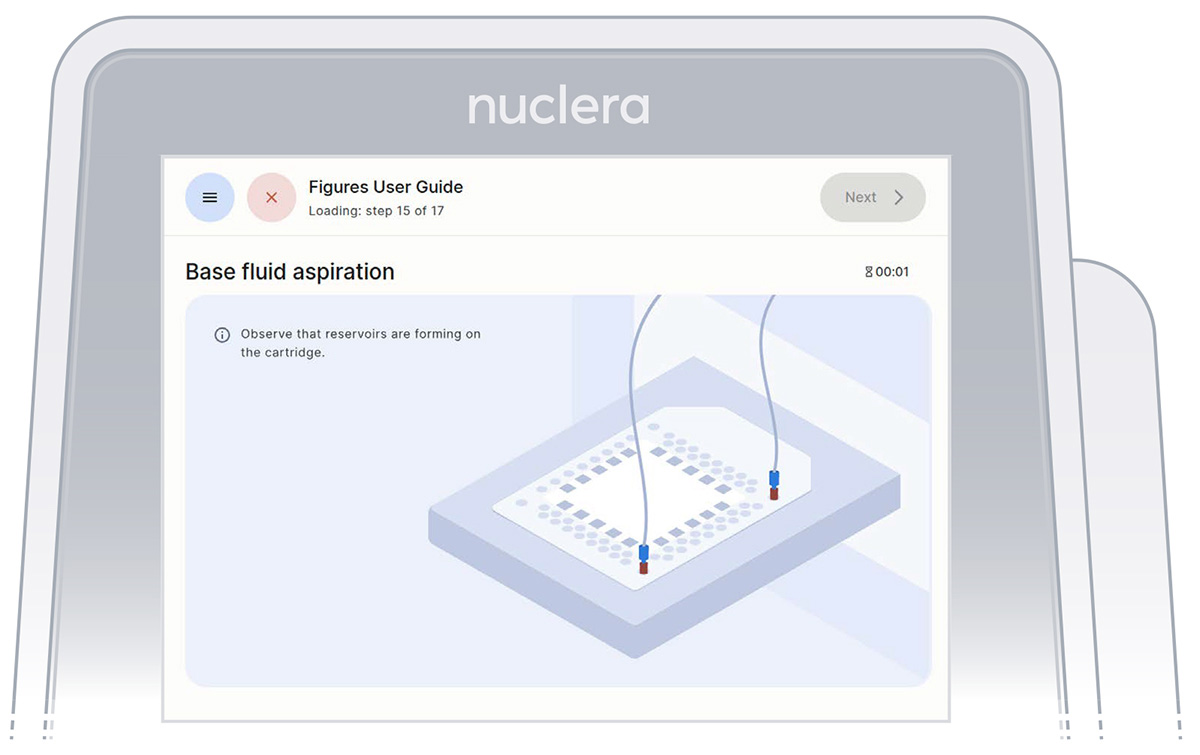 Figure 26: Base fluid aspiration
Figure 26: Base fluid aspiration
- Disconnect the tubes from the cartridge and place them in the tube holder on the right hand side of the instrument. Press the [Next] button on the screen (Figure 27), and the drawer will close. Loading checks will be performed, and the drawer will open.
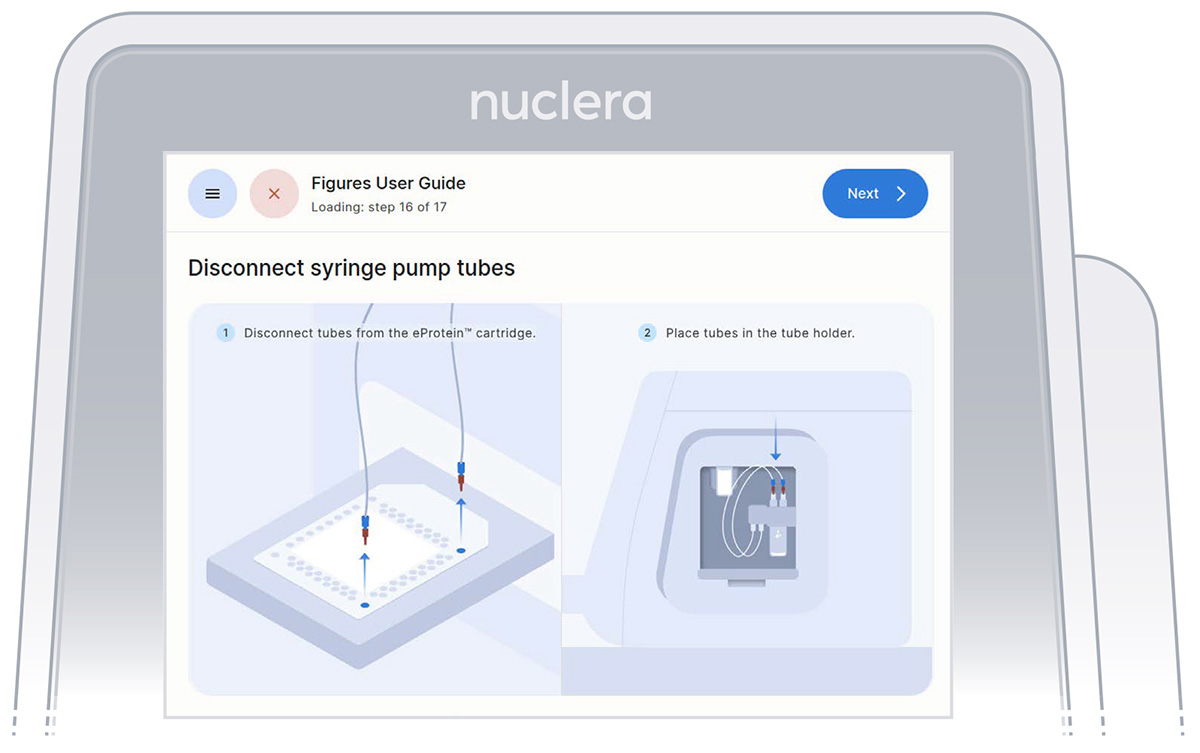 Figure 27: Disconnect the tubes and place them on the tube holder
Figure 27: Disconnect the tubes and place them on the tube holder
- Inspect the reservoirs have formed correctly in the cartridge as shown on the screen. If so, Press the [Next] button (Figure 28).
Note: Any presence of a marker on the reservoir is acceptable, as shown on the top right of Figure 29
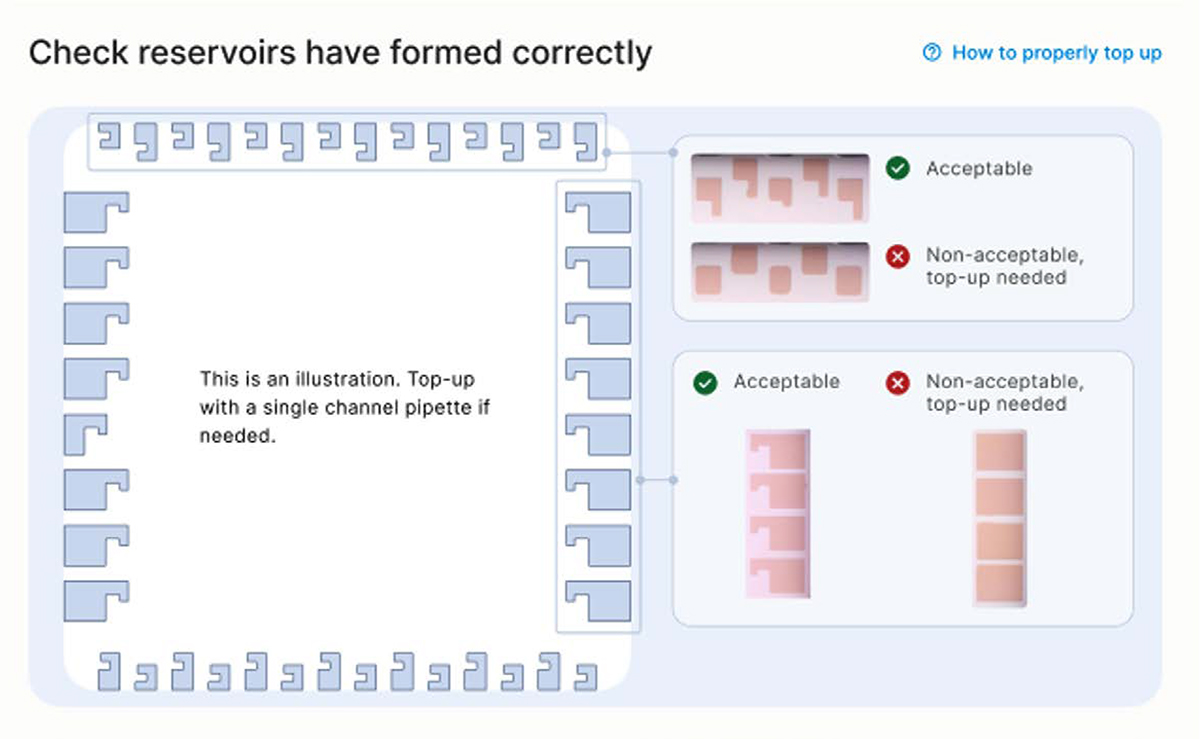 Figure 28: Check the reservoirs have formed correctly on the cartridge
Figure 28: Check the reservoirs have formed correctly on the cartridge
Troubleshooting tip 1:
If a reservoir is not properly formed, first fully engage a new empty pipette tip into the port and reach the bottom of the port. This action may trigger the correct formation of the reservoir.
Troubleshooting tip 2:
▷ If the shape of a reservoir is still not correct, remove the empty tip from the port, replace with a new tip, then add a small volume of the corresponding reagent using a p20 pipette with a pipette tip. Do not depress the pipette past the first stop as this could introduce air bubbles inside the cartridge.
▷ Re-engage the tip until reaching the bottom to the port and dispense the reagent slowly until correction is complete (Figure 29).
▷ The recommended volumes for manual correction are:
▷ 1.5 µL for ports in rows A, B, C or H
▷ 3 µL for ports in columns 10 or 12
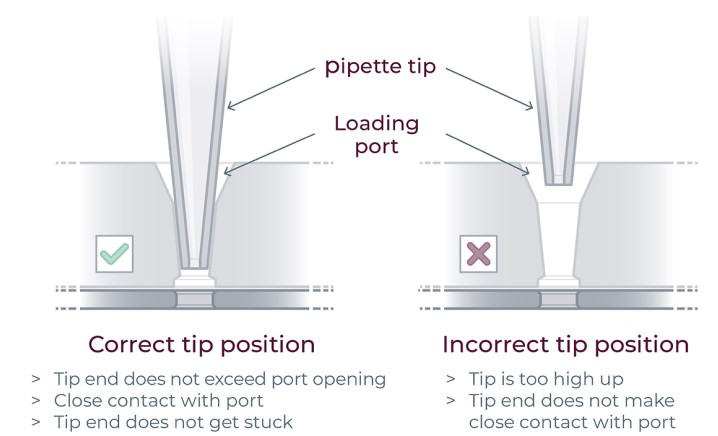
Figure 29: For manual correction of the reagent loading to correctly form the reservoirs on cartridge
Once the experiment starts, it will take approximately 24 hours to complete. You can monitor its progress on the screen.
Analyze the results
Instrument software results screen
After completion of the experiment, the results are shown on the instrument screen. The four best obtainable combinations of eGene and Cell-free Blend are displayed with the predicted in-tube scale-up yields (Figure 30).
 Figure 30: Result screen from the Instrument Software
Figure 30: Result screen from the Instrument Software
eProtein Discovery report
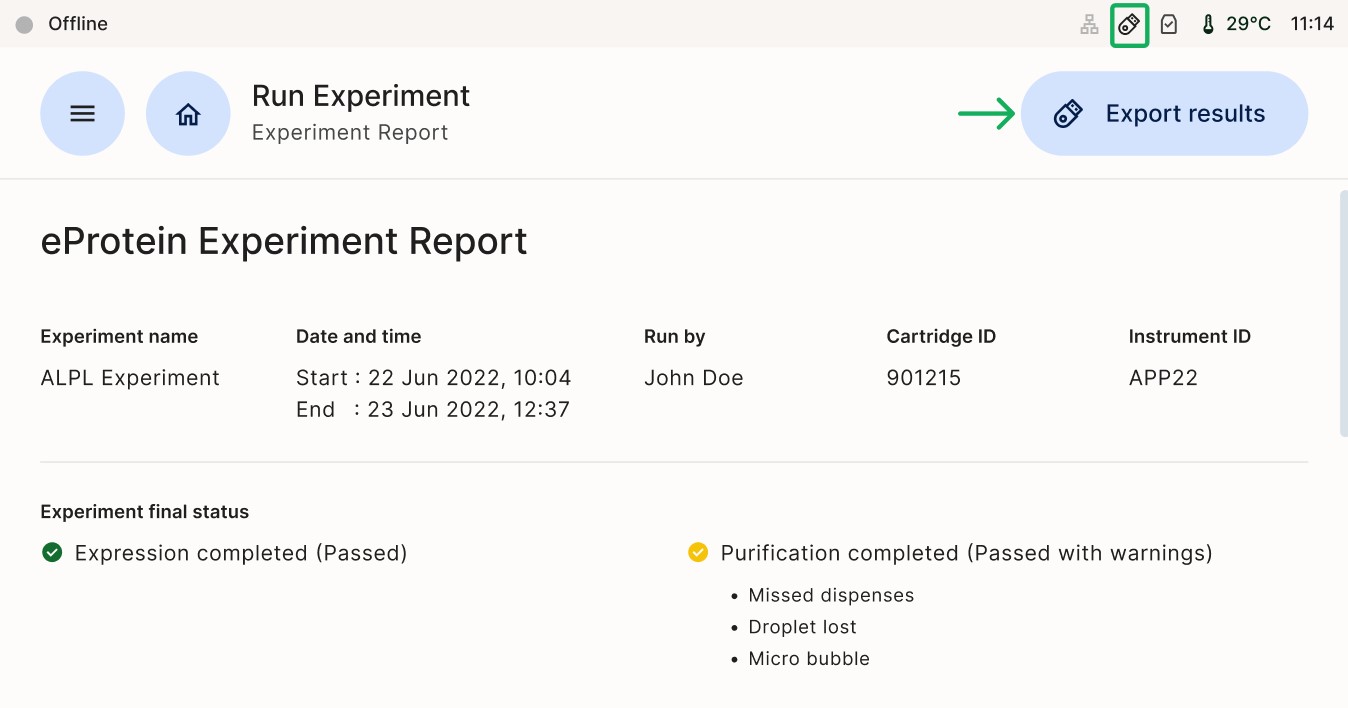
At the end of the experiment a report containing all the information about the experimental setup and the results can be downloaded from the instrument. Data can be exported using a USB, LAN or a Local Laptop. The upload takes about 15 minutes and during this time the [Download Report] button at the top right corner of the screen is grayed out.
USB
To export data to a USB flash drive it required to have a company-approved USB flash drive for data transfer. Encrypted flash drives are not currently supported.
Insert the flash drive on the right hand side of the instrument in the USB port. This will highlight the flashdrive icon on the screen.
Click on [Export Results]
A selection Window will show the two options available. Select (a) USB drive and click on (b) Export in the next window as shown in figure below.
(a)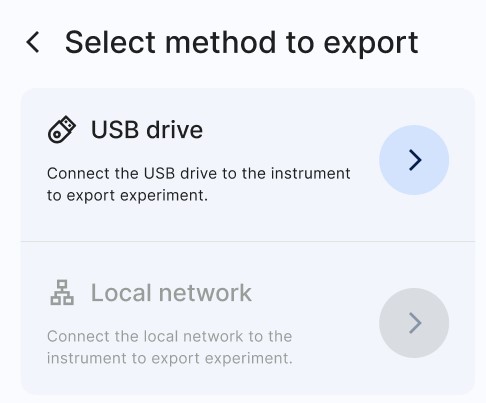 (b)
(b)
Data is retrieved from the instrument by inserting a USB flash drive into the instrument and exporting the data onto it, then inserting it into a Windows laptop and using an Excel spreadsheet to analyze the data. For data visualisation and analysis, it is required to have a laptop with Microsoft Excel
LAN and Local Laptop
Connect an Ethernet cable directly from the instrument's Ethernet port to the Windows laptop (or to the Ethernet-to-USB dongle).
On the instrument:
Check that the LAN icon is no longer greyed-out.
Open a completed experiment and tap the “Export results” button.
Tap (a) "Local network" and enter the following (b):
URL: smb://192.168.1.100/eProteinReports
Username: the name of the Windows user that has access to the shared directory, if this is a domain user then the username should be of the form DOMAINNAME\username
Password: the password of the Windows user that has access to the shared directory
(a)  (b)
(b) 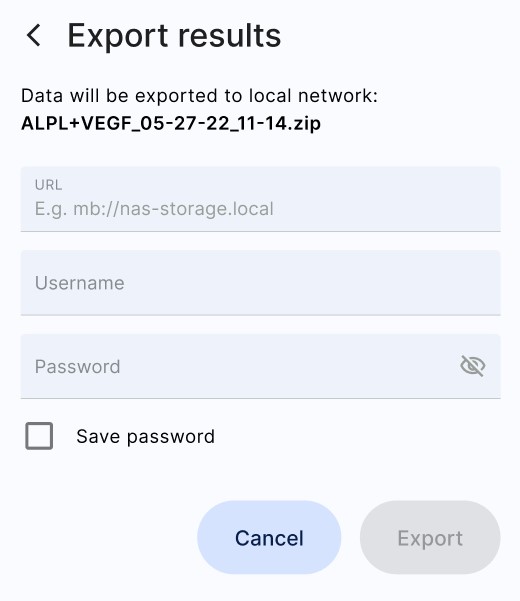
Note: the instrument should not be switched off until the report is transferred.
The experiment report contains:
▷ Experiment video
The experiment video provides a record of cartridge droplet operation, allowing users to review performance for quality control, troubleshoot issues, or verify specific events during the run. Any questions or concerns regarding the operation of the droplets should be directed to the Nuclera Technical Support team (techsupport@nuclera.com).
▷ PDF report file
The PDF report file is a summary of the experiment setup and the results, saved in the report folder with the name given to the experiment included in the file name
▷ CSV report file The report file is a csv file saved in the report folder with the name given to the experiment included in the file name. The results for each one of the 30 purified target protein conditions, and the 192 conditions for the produced protein are listed in the csv file. It also contains the measured values for the controls, the expected range for the controls, and a PASS/FAIL score if the measured values are within the expected range.
▷ Blue light images (TIFF images)
Images acquired at the end of expressions and purification. These images can give the user
information about the solubility of the protein.
▷ Other files
The folder contains additional files that can be used by the Nuclera Technical Support team for troubleshooting purpose.
Visualise your data
- Unzip (right click + Extract All) your report folder exported from eProtein Discovery instrument
- Open the report.csv file and select the entire sheet by clicking the top left corner, or by using the shortcut "Ctrl + A" or "Cmd + A".
- Copy the data using the shortcut "Ctrl + C" or "Cmd + C".
- Paste the report in tab "3. Paste Report Here" of the eProtein Discovery Standalone Template (compatible with Microsoft Excel, not compatible with Google Sheet).
- Review the data in the "4. Output" tab and ensure it is correctly labelled with corresponding constructs and cell-free blend labels.
- The labelled data charts will be automatically plotted in the "5. Charts" tab ready for your review.
Finishing the experiment
- Remove the cartridge from the instrument drawer by lifting it as shown on the screen and place it in its original packaging. Press the [Next] button (Figure 31).
Note: there is no need to drain the base fluid out of the cartridge.
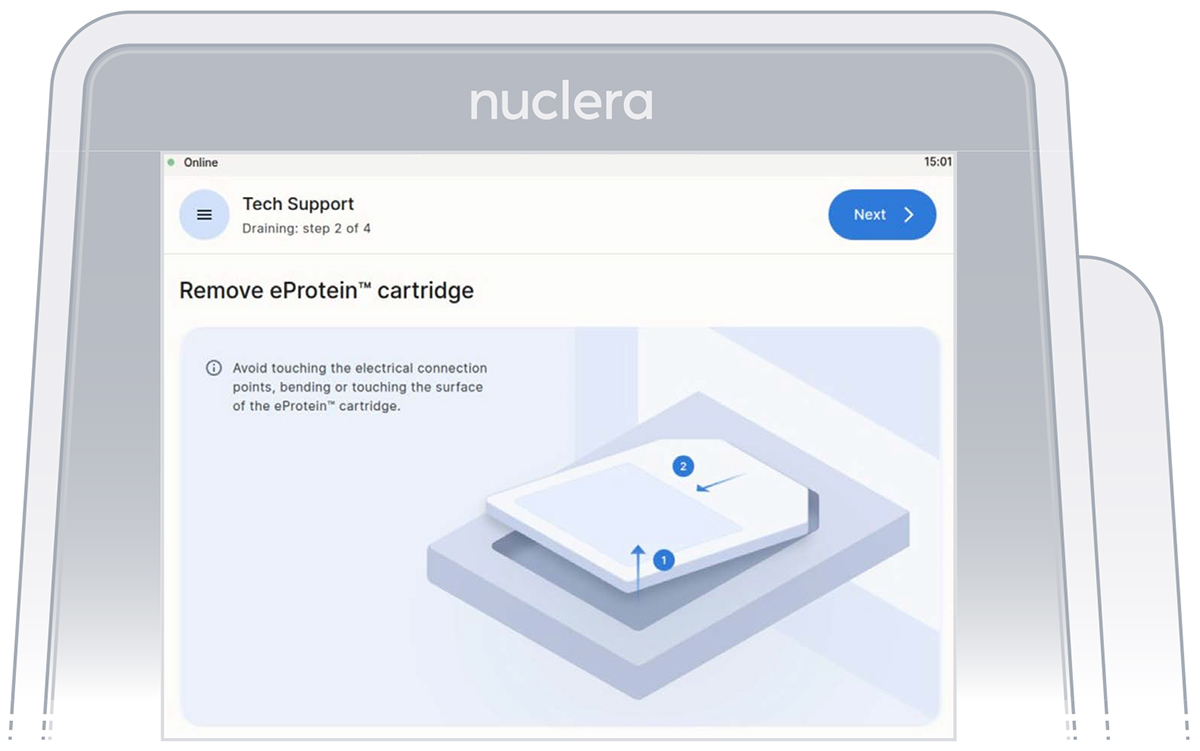
- Remove the waste container from the holder, empty its content, and place it back on the instrument.
- Remove the vial of base fluid and dispose of it with biohazard sharps waste container according to local waste disposal rules and regulations.
Note: Do not reuse consumed cartridges and dispose of any residual reagents, kits are intended as single use only.
- Dispose the packaged used cartridge in a biohazard sharps container, according to local waste disposal rules and regulations.
- The experiment report is available for download from the eProtein Discovery software.
- Power down the instrument after use by pressing the [Power off] button (Figure 32)
Frequently Asked Questions (FAQ)
| Questions/Issues | Answers |
|---|---|
| How do I contact the support team? | Email us at techsupport@nuclera.com Call us at +441223942 761 (UK phone number / WhatsApp Business) or +1 508-306-1297 (US phone number) |
| Where can I suggest future improvements? | Please email us at techsupport@nuclera.com, your feedback is very important to us as it allows us to improve the instrument, the technology and our services. |
| A component or reagent is missing | If a component or a reagent is missing, please contact the Nuclera Technical Support team |
Nuclera Technical Support:
UK Phone: +44 1223 942 761
US Phone: +1 508-306-1297
Email: techsupport@nuclera.com
Offices:
Nuclera UK (HQ):
One Vision Park, Station Road, Cambridge, CB24 9NP, UK
Nuclera USA:
1000 Technology Park Drive, Suite B, Billerica MA 01821, USA
www.nuclera.com
Copyright © 2025 Nuclera Ltd. All trademarks are the property of Nuclera, Ltd. Visit nuclera.com/legal for more info.