Assembly Guidelines to convert Linear eGene™ constructs into plasmids
This protocol provides general guidelines to clone linear eGene™ constructs into an expression vector of choice using Gibson Assembly® for use in E. coli scale up. The guidelines include primer design considerations supporting insert amplification, vector linearization, Gibson Assembly®, and transformation
Reagents and Materials required
| Item | Item |
|---|---|
| Desired eGene™ construct | DpnI restriction endonuclease |
| Desired expression vector | PCR clean up kit |
| Appropriate primers to amplify eGene insert (see Guidelines) | Gibson Assembly® Master Mix (commercial or homemade) |
| Appropriate primers to amplify vector (see Guidelines) | DNA quantification tool (e.g. Nanodrop® or Qubit®) |
| Nuclease-free water | Competent cells (for transformation) |
| High-fidelity polymerase PCR master mix | LB agar plate with appropriate antibiotic |
| PCR tubes | LB medium |
| Thermocycler | Antibiotic appropriate for plasmid |
The transition from an eProtein Discovery™ screen to E. coli expression has only been validated using the pET vector series. Any alterations—such as changes to the promoter, removal of the lac operator/inducer, or differences in the ribosome binding site (RBS) or its spacing from the promoter may impact expression success.
Guidelines
Primer design
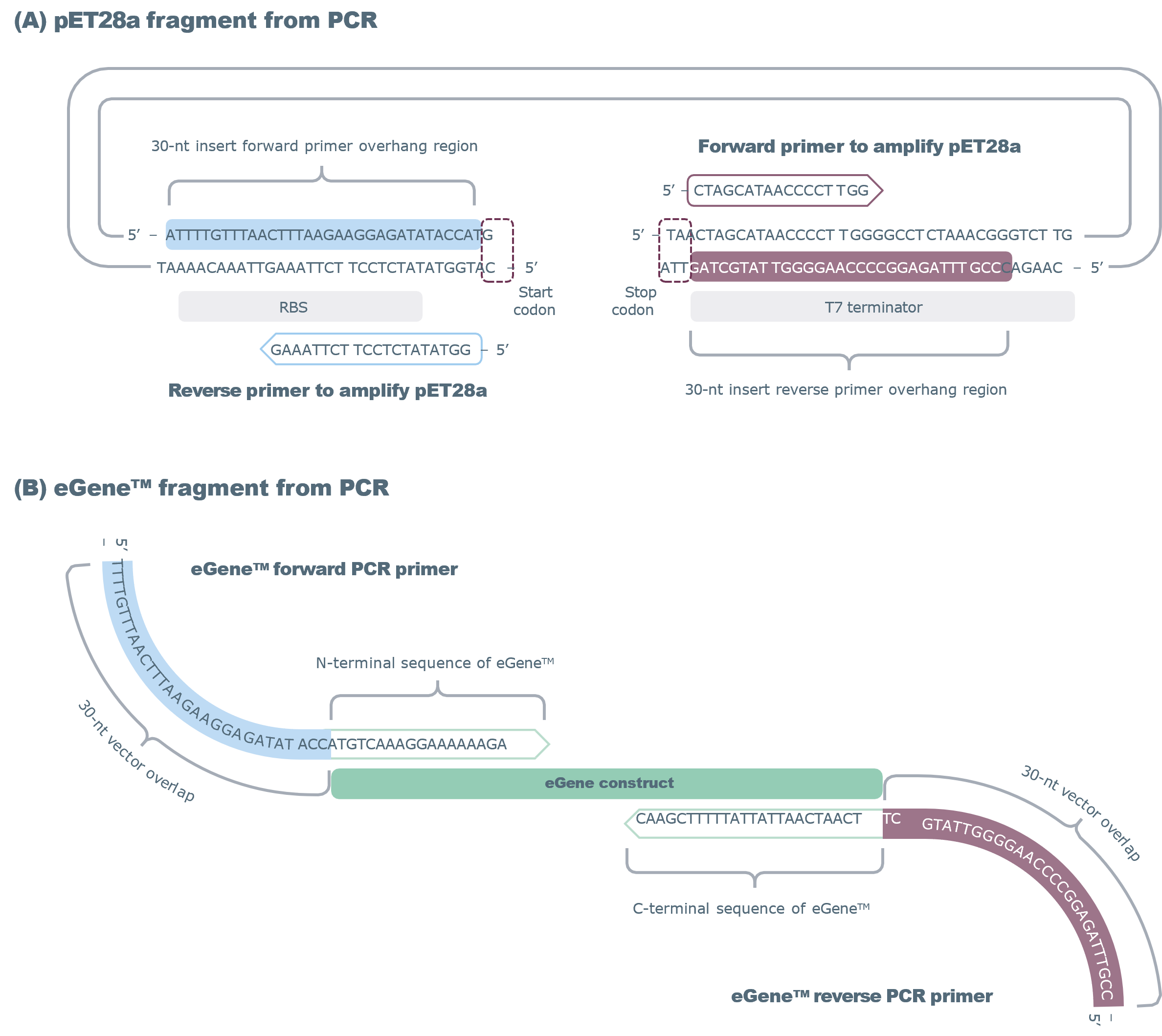
For Gibson Assembly®, consider the vector and insert as two fragments to be joined seamlessly into a circular plasmid. Therefore, two sets of primers (and two PCR reactions) will be required to amplify the expression vector and eGene™ selected. Below are guidelines for designing primers to amplify the vector and eGene™ insert. See Figure 1 for an example with pET28a.
Amplifying the Vector (Inverse PCR)
- Design the reverse primer to begin immediately upstream of ATG START codon (scan for the START codon just downstream of the Shine Dalgarno sequence/RBS). The 5’ end should not include CAT (reverse complement of ATG).
- Design forward primer to begin immediately after the TAA/TAG/TGA stop codon (just upstream of the terminator). The 5’ end should not include the stop codon.
- Primers should be:
- 18–30 bp in length.
- Annealing temperature should be between 55 and 70°C.
- Ensure Tm difference between primers is ≤ 5°C; both should anneal below the extension temperature of your polymerase.
- Design primers with 40–60% GC content (ideal: ~50–55%).
- Recommended: Use GC clamps (G/C at 3′ ends), avoid secondary structures, and calculate Tm using tools like NEBuilder® or IDT OligoAnalyzer®.
- Avoid self-complementarity within the primers and between the primer pair.
Amplifying the eGene™ Insert
- Design forward and reverse primers with 30 bp 5′ overhangs complementary to the ends of your linearized vector.
- Append the 30 bp overhang sequences to eGene™-specific primers using the loci-specific sequences provided in Table 2.
| Primer | Sequence (5’ to 3’) |
|---|---|
| Forward | **[30 bp vector overhang]**ATGTCAAAGGAAAAAAGA |
| Reverse | **[30 bp vector overhang]**TCAATCAATTATTATTTTTCGAAC |
General considerations for PCR
-
Use a high-fidelity PCR Master Mix for both vector and insert amplification.
-
Follow the manufacturer’s guidelines for PCR Master Mix preparation and thermocycler settings.
-
Amplifying the vector (Inverse PCR):
- To prevent false positives during transformation, digest the original plasmid template using DpnI. After PCR amplification, add 20 units of DpnI per 50 μL of PCR product, and incubate at 37°C for 30 minutes. Heat-inactivate the enzyme by incubating at 80°C for 20 minutes before proceeding with clean-up.
- Prior to PCR purification, check the PCR product on a 1% agarose gel. If you observe a single band of the expected size, proceed to purify the PCR product using any PCR cleanup method of your choice.
- Alternatively, resolve the bands on an agarose gel and proceed to excise the correct band to purify using gel extraction kit.
-
Amplifying the eGene™ Insert:
- When amplifying the eGene™ insert the elongation time should take into consideration the full length of the eGene, including the gene of interest (GOI). See Table 3 for lengths.
- Visualize the PCR product on a 1% agarose gel. If single bands are visible, proceed with PCR clean-up. If bands are faint or diffuse, use the band stab protocol from the eGene Prep guide. Do not use gel extraction for eGene™ inserts, as this may lower yields.
- PCR clean up can be performed using any PCR cleanup method of your choice.
| eGene™ insert solubility tag variant | Size of insert |
|---|---|
| P17 | 386 bp + bp of GOI |
| CUSF | 554 bp + bp of GOI |
| FH8 | 491 bp + bp of GOI |
| TRX | 611 bp + bp of GOI |
| ZZ | 635 bp + bp of GOI |
| SUMO | 593 bp + bp of GOI |
| SNUT | 728 bp + bp of GOI |
| No solubility tag | 242 bp + bp of GOI |
General considerations for Gibson Assembly® and transformation
- For Gibson Assembly® follow the manufacturer’s instructions to assemble the insert and the linearized vector.
- It is recommended to transform the assembled product into competent E. coli (e.g., NEB® 5-alpha) and plate in LB agar plates containing the relevant antibiotic for the plasmid and incubate overnight at 37°C.
- The linearized vector can be used as a transformation negative control. Ideally you should not see any colonies in this plate.
- (Optional) Select 5 random colonies, from the plates incubated overnight, to verify the presence of the insert by colony PCR (cPCR) using cPCR forward and reverse primer. cPCR forward and reverse primers can be found in Table 4.
| Primer | Sequence (5’ to 3’) |
|---|---|
| Forward | ATGTCAAAGGAAAAAAGA |
| Reverse | TCAATCAATTATTATTTTTCGAAC |
- Inoculate antibiotic supplemented medium relevant to the plasmid with 3-5 cPCR confirmed colonies for miniprep and subsequent sequence analysis.
- Transform sequence verified plasmids into an E. coli expression strain of choice.
Disclaimers: NanoDrop and Qubit are registered trademarks of Thermo Fisher Scientific. IDT OligoAnalyzer is a registered trademark of IDT. Gibson Assembly is a registered trademark of Telesis Bio Inc. NEBuilder and NEB are registered trademarks of New England Biolabs.
Nuclera Technical Support:
UK Phone +44 1223 942 761
US Phone: +1 508-306-1297
Email: techsupport@nuclera.com
Offices:
Nuclera UK (HQ):
One Vision Park, Station Road, Cambridge, CB24 9NP, UK
Nuclera USA: 1000 Technology Park Drive, Suite B, Billerica MA 01821, USA www.nuclera.com
Copyright © 2025 Nuclera Ltd. All trademarks are the property of Nuclera, Ltd. Visit nuclera.com/legal for more info.